User login
A massive collaborative project spanning four continents and 744 research centers has revealed driver mutations in both protein-coding and noncoding regions of 38 cancer types.
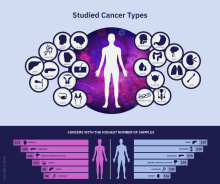
The Pan-Cancer Analysis of Whole Genomes (PCAWG) is an integrative analysis of the whole-genome sequences from 2,658 donors across 38 common tumor types. The findings are expected to add exponentially to what’s currently known about the complex genetics of cancer, and they point to possible strategies for improving cancer prevention, diagnosis, and care.
Six articles summarizing the findings are presented in a series of papers in Nature, and 16 more appear in affiliated publications.
“It’s humbling that it was only 14 years ago that the genomics community sequenced its very first cancer exome, and it was able to identify mutations within the roughly 20,000 protein-coding genes in the human cell,” investigator Lincoln Stein, MD, PhD, of the Ontario Institute for Cancer Research in Toronto, said in a telephone briefing.
Exome sequencing, however, covers only protein-coding genomic regions, which constitute only about 1% of the entire genome, “so assembling an accurate portrait of the cancer genome using just the exome data is like trying to put together a 100,000-piece jigsaw puzzle when you’re missing 99% of the pieces and there’s no puzzle box with a completed picture to guide you,” Dr. Stein said.
Members of the PCAWG from centers in North America, Europe, Asia, and Australia screened 2,658 whole-cancer genomes and matched samples of noncancerous tissues from the same individuals, along with 1,188 transcriptomes cataloging the sequences and expression of RNA transcripts in a given tumor. The 6-year project netted more than 800 terabytes of genomic data, roughly equivalent to the digital holdings of the U.S. Library of Congress multiplied by 11.
The findings are summarized in papers focusing on cancer drivers, noncoding changes, mutational signatures, structural variants, cancer evolution over time, and RNA alterations.
Driver mutations
Investigators found that the average cancer genome contains four or five driver mutations located in both coding and noncoding regions. They also found, however, that in approximately 5% of cases no driver mutations could be identified.
A substantial proportion of tumors displayed “hallmarks of genomic catastrophes.” About 22% of tumors exhibited chromothripsis, a mutational process marked by hundreds or even thousands of clustered chromosomal rearrangements. About 18% showed chromoplexy, which is characterized by scattering and rearrangement of multiple strands of DNA from one or more chromosomes.
Analyzing driver point mutations and structural variants in noncoding regions, the investigators found the usual suspects – previously reported culprits – as well as novel candidates.
For example, they identified point mutations in the five prime region of the tumor suppressor gene TP53 and the three prime untranslated regions of NFKBIZ (a nuclear factor kappa B inhibitor) and TOB1 (an antiproliferative protein), focal deletion in BRD4 (a transcriptional and epigenetic regulator), and rearrangements in chromosomal loci in members of the AKR1C family of enzymes thought to play a role in disease progression.
In addition, investigators identified mutations in noncoding regions of TERT, a telomerase gene. These mutations result in ramped-up expression of telomerase, which in turn promotes uncontrollable division of tumor cells.
Mutational signatures
In a related line of research, PCAWG investigators identified new DNA mutational signatures ranging from single nucleotide polymorphisms to insertions and deletions, as well as to structural variants – rearrangements of large sections of the genome.
“The substantial size of our dataset, compared with previous analyses, enabled the discovery of new signatures, the separation of overlapping signatures, and the decomposition of signatures into components that may represent associated – but distinct – DNA damage, repair, and/or replication mechanisms. By estimating the contribution of each signature to the mutational catalogs of individual cancer genomes, we revealed associations of signatures to exogenous or endogenous exposures, as well as to defective DNA maintenance processes,” the investigators wrote.
They also acknowledged, however, that “many signatures are of unknown cause.”
Cancer evolution
One of the six main studies focused on the evolution of cancer over time. Instead of providing a “snapshot” of the genome as captured by sequencing tissue from a single biopsy, consortium investigators created full-length features of the “life history and evolution of mutational processes and driver mutation sequences.”
They found that early cancer development was marked by relatively few mutations in driver genes and by identifiable copy-number gains, including trisomy 7 in glioblastoma, and an abnormal mirroring of the arms (isochromosome) of chromosome 17 in medulloblastoma.
In 40% of the samples, however, there were significant changes in the mutational spectrum as the cancers grew, leading to a near quadrupling of driver genes and increased genomic instability in later-stage tumors.
“Copy-number alterations often occur in mitotic crises and lead to simultaneous gains of chromosomal segments,” the investigators wrote. “Timing analyses suggest that driver mutations often precede diagnosis by many years, if not decades. Together, these results determine the evolutionary trajectories of cancer and highlight opportunities for early cancer detection.”
Implications for cancer care
“When I used to treat patients with cancer, I was always completely amazed and puzzled by how two patients could have what looked like the same tumor. It would look the same under the microscope, have the same size, and the two patients would receive exactly the same treatment, but the two patients would have completely opposite outcomes; one would survive, and one would die. What this analysis … has done is really laid bare the reasons for that unpredictability in clinical outcomes,” Peter Campbell, MD, PhD, of the Wellcome Sanger Institute in Hinxton, England, said during the telebriefing.
“The most striking finding out of all of the suite of papers is just how different one person’s cancer genome is from another person’s. We see thousands of different combinations of mutations that can cause the cancer, and more than 80 different underlying processes generating the mutations in a cancer, and that leads to very different shapes and patterns in the genome that result,” he added.
On a positive note, the research shows that one or more driver mutations can be identified in about 95% of all cancer patients, and it elucidates the sequence of events leading to oncogenesis and tumor evolution, providing opportunities for earlier identification and potential interventions to prevent cancer, Dr. Campbell said.
The PCAWG was a collaborative multinational effort with multiple funding sources and many investigators.
SOURCE: Nature. 2020 Feb 5. https://www.nature.com/collections/pcawg/
A massive collaborative project spanning four continents and 744 research centers has revealed driver mutations in both protein-coding and noncoding regions of 38 cancer types.
The Pan-Cancer Analysis of Whole Genomes (PCAWG) is an integrative analysis of the whole-genome sequences from 2,658 donors across 38 common tumor types. The findings are expected to add exponentially to what’s currently known about the complex genetics of cancer, and they point to possible strategies for improving cancer prevention, diagnosis, and care.
Six articles summarizing the findings are presented in a series of papers in Nature, and 16 more appear in affiliated publications.
“It’s humbling that it was only 14 years ago that the genomics community sequenced its very first cancer exome, and it was able to identify mutations within the roughly 20,000 protein-coding genes in the human cell,” investigator Lincoln Stein, MD, PhD, of the Ontario Institute for Cancer Research in Toronto, said in a telephone briefing.
Exome sequencing, however, covers only protein-coding genomic regions, which constitute only about 1% of the entire genome, “so assembling an accurate portrait of the cancer genome using just the exome data is like trying to put together a 100,000-piece jigsaw puzzle when you’re missing 99% of the pieces and there’s no puzzle box with a completed picture to guide you,” Dr. Stein said.
Members of the PCAWG from centers in North America, Europe, Asia, and Australia screened 2,658 whole-cancer genomes and matched samples of noncancerous tissues from the same individuals, along with 1,188 transcriptomes cataloging the sequences and expression of RNA transcripts in a given tumor. The 6-year project netted more than 800 terabytes of genomic data, roughly equivalent to the digital holdings of the U.S. Library of Congress multiplied by 11.
The findings are summarized in papers focusing on cancer drivers, noncoding changes, mutational signatures, structural variants, cancer evolution over time, and RNA alterations.
Driver mutations
Investigators found that the average cancer genome contains four or five driver mutations located in both coding and noncoding regions. They also found, however, that in approximately 5% of cases no driver mutations could be identified.
A substantial proportion of tumors displayed “hallmarks of genomic catastrophes.” About 22% of tumors exhibited chromothripsis, a mutational process marked by hundreds or even thousands of clustered chromosomal rearrangements. About 18% showed chromoplexy, which is characterized by scattering and rearrangement of multiple strands of DNA from one or more chromosomes.
Analyzing driver point mutations and structural variants in noncoding regions, the investigators found the usual suspects – previously reported culprits – as well as novel candidates.
For example, they identified point mutations in the five prime region of the tumor suppressor gene TP53 and the three prime untranslated regions of NFKBIZ (a nuclear factor kappa B inhibitor) and TOB1 (an antiproliferative protein), focal deletion in BRD4 (a transcriptional and epigenetic regulator), and rearrangements in chromosomal loci in members of the AKR1C family of enzymes thought to play a role in disease progression.
In addition, investigators identified mutations in noncoding regions of TERT, a telomerase gene. These mutations result in ramped-up expression of telomerase, which in turn promotes uncontrollable division of tumor cells.
Mutational signatures
In a related line of research, PCAWG investigators identified new DNA mutational signatures ranging from single nucleotide polymorphisms to insertions and deletions, as well as to structural variants – rearrangements of large sections of the genome.
“The substantial size of our dataset, compared with previous analyses, enabled the discovery of new signatures, the separation of overlapping signatures, and the decomposition of signatures into components that may represent associated – but distinct – DNA damage, repair, and/or replication mechanisms. By estimating the contribution of each signature to the mutational catalogs of individual cancer genomes, we revealed associations of signatures to exogenous or endogenous exposures, as well as to defective DNA maintenance processes,” the investigators wrote.
They also acknowledged, however, that “many signatures are of unknown cause.”
Cancer evolution
One of the six main studies focused on the evolution of cancer over time. Instead of providing a “snapshot” of the genome as captured by sequencing tissue from a single biopsy, consortium investigators created full-length features of the “life history and evolution of mutational processes and driver mutation sequences.”
They found that early cancer development was marked by relatively few mutations in driver genes and by identifiable copy-number gains, including trisomy 7 in glioblastoma, and an abnormal mirroring of the arms (isochromosome) of chromosome 17 in medulloblastoma.
In 40% of the samples, however, there were significant changes in the mutational spectrum as the cancers grew, leading to a near quadrupling of driver genes and increased genomic instability in later-stage tumors.
“Copy-number alterations often occur in mitotic crises and lead to simultaneous gains of chromosomal segments,” the investigators wrote. “Timing analyses suggest that driver mutations often precede diagnosis by many years, if not decades. Together, these results determine the evolutionary trajectories of cancer and highlight opportunities for early cancer detection.”
Implications for cancer care
“When I used to treat patients with cancer, I was always completely amazed and puzzled by how two patients could have what looked like the same tumor. It would look the same under the microscope, have the same size, and the two patients would receive exactly the same treatment, but the two patients would have completely opposite outcomes; one would survive, and one would die. What this analysis … has done is really laid bare the reasons for that unpredictability in clinical outcomes,” Peter Campbell, MD, PhD, of the Wellcome Sanger Institute in Hinxton, England, said during the telebriefing.
“The most striking finding out of all of the suite of papers is just how different one person’s cancer genome is from another person’s. We see thousands of different combinations of mutations that can cause the cancer, and more than 80 different underlying processes generating the mutations in a cancer, and that leads to very different shapes and patterns in the genome that result,” he added.
On a positive note, the research shows that one or more driver mutations can be identified in about 95% of all cancer patients, and it elucidates the sequence of events leading to oncogenesis and tumor evolution, providing opportunities for earlier identification and potential interventions to prevent cancer, Dr. Campbell said.
The PCAWG was a collaborative multinational effort with multiple funding sources and many investigators.
SOURCE: Nature. 2020 Feb 5. https://www.nature.com/collections/pcawg/
A massive collaborative project spanning four continents and 744 research centers has revealed driver mutations in both protein-coding and noncoding regions of 38 cancer types.
The Pan-Cancer Analysis of Whole Genomes (PCAWG) is an integrative analysis of the whole-genome sequences from 2,658 donors across 38 common tumor types. The findings are expected to add exponentially to what’s currently known about the complex genetics of cancer, and they point to possible strategies for improving cancer prevention, diagnosis, and care.
Six articles summarizing the findings are presented in a series of papers in Nature, and 16 more appear in affiliated publications.
“It’s humbling that it was only 14 years ago that the genomics community sequenced its very first cancer exome, and it was able to identify mutations within the roughly 20,000 protein-coding genes in the human cell,” investigator Lincoln Stein, MD, PhD, of the Ontario Institute for Cancer Research in Toronto, said in a telephone briefing.
Exome sequencing, however, covers only protein-coding genomic regions, which constitute only about 1% of the entire genome, “so assembling an accurate portrait of the cancer genome using just the exome data is like trying to put together a 100,000-piece jigsaw puzzle when you’re missing 99% of the pieces and there’s no puzzle box with a completed picture to guide you,” Dr. Stein said.
Members of the PCAWG from centers in North America, Europe, Asia, and Australia screened 2,658 whole-cancer genomes and matched samples of noncancerous tissues from the same individuals, along with 1,188 transcriptomes cataloging the sequences and expression of RNA transcripts in a given tumor. The 6-year project netted more than 800 terabytes of genomic data, roughly equivalent to the digital holdings of the U.S. Library of Congress multiplied by 11.
The findings are summarized in papers focusing on cancer drivers, noncoding changes, mutational signatures, structural variants, cancer evolution over time, and RNA alterations.
Driver mutations
Investigators found that the average cancer genome contains four or five driver mutations located in both coding and noncoding regions. They also found, however, that in approximately 5% of cases no driver mutations could be identified.
A substantial proportion of tumors displayed “hallmarks of genomic catastrophes.” About 22% of tumors exhibited chromothripsis, a mutational process marked by hundreds or even thousands of clustered chromosomal rearrangements. About 18% showed chromoplexy, which is characterized by scattering and rearrangement of multiple strands of DNA from one or more chromosomes.
Analyzing driver point mutations and structural variants in noncoding regions, the investigators found the usual suspects – previously reported culprits – as well as novel candidates.
For example, they identified point mutations in the five prime region of the tumor suppressor gene TP53 and the three prime untranslated regions of NFKBIZ (a nuclear factor kappa B inhibitor) and TOB1 (an antiproliferative protein), focal deletion in BRD4 (a transcriptional and epigenetic regulator), and rearrangements in chromosomal loci in members of the AKR1C family of enzymes thought to play a role in disease progression.
In addition, investigators identified mutations in noncoding regions of TERT, a telomerase gene. These mutations result in ramped-up expression of telomerase, which in turn promotes uncontrollable division of tumor cells.
Mutational signatures
In a related line of research, PCAWG investigators identified new DNA mutational signatures ranging from single nucleotide polymorphisms to insertions and deletions, as well as to structural variants – rearrangements of large sections of the genome.
“The substantial size of our dataset, compared with previous analyses, enabled the discovery of new signatures, the separation of overlapping signatures, and the decomposition of signatures into components that may represent associated – but distinct – DNA damage, repair, and/or replication mechanisms. By estimating the contribution of each signature to the mutational catalogs of individual cancer genomes, we revealed associations of signatures to exogenous or endogenous exposures, as well as to defective DNA maintenance processes,” the investigators wrote.
They also acknowledged, however, that “many signatures are of unknown cause.”
Cancer evolution
One of the six main studies focused on the evolution of cancer over time. Instead of providing a “snapshot” of the genome as captured by sequencing tissue from a single biopsy, consortium investigators created full-length features of the “life history and evolution of mutational processes and driver mutation sequences.”
They found that early cancer development was marked by relatively few mutations in driver genes and by identifiable copy-number gains, including trisomy 7 in glioblastoma, and an abnormal mirroring of the arms (isochromosome) of chromosome 17 in medulloblastoma.
In 40% of the samples, however, there were significant changes in the mutational spectrum as the cancers grew, leading to a near quadrupling of driver genes and increased genomic instability in later-stage tumors.
“Copy-number alterations often occur in mitotic crises and lead to simultaneous gains of chromosomal segments,” the investigators wrote. “Timing analyses suggest that driver mutations often precede diagnosis by many years, if not decades. Together, these results determine the evolutionary trajectories of cancer and highlight opportunities for early cancer detection.”
Implications for cancer care
“When I used to treat patients with cancer, I was always completely amazed and puzzled by how two patients could have what looked like the same tumor. It would look the same under the microscope, have the same size, and the two patients would receive exactly the same treatment, but the two patients would have completely opposite outcomes; one would survive, and one would die. What this analysis … has done is really laid bare the reasons for that unpredictability in clinical outcomes,” Peter Campbell, MD, PhD, of the Wellcome Sanger Institute in Hinxton, England, said during the telebriefing.
“The most striking finding out of all of the suite of papers is just how different one person’s cancer genome is from another person’s. We see thousands of different combinations of mutations that can cause the cancer, and more than 80 different underlying processes generating the mutations in a cancer, and that leads to very different shapes and patterns in the genome that result,” he added.
On a positive note, the research shows that one or more driver mutations can be identified in about 95% of all cancer patients, and it elucidates the sequence of events leading to oncogenesis and tumor evolution, providing opportunities for earlier identification and potential interventions to prevent cancer, Dr. Campbell said.
The PCAWG was a collaborative multinational effort with multiple funding sources and many investigators.
SOURCE: Nature. 2020 Feb 5. https://www.nature.com/collections/pcawg/
FROM NATURE