User login
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.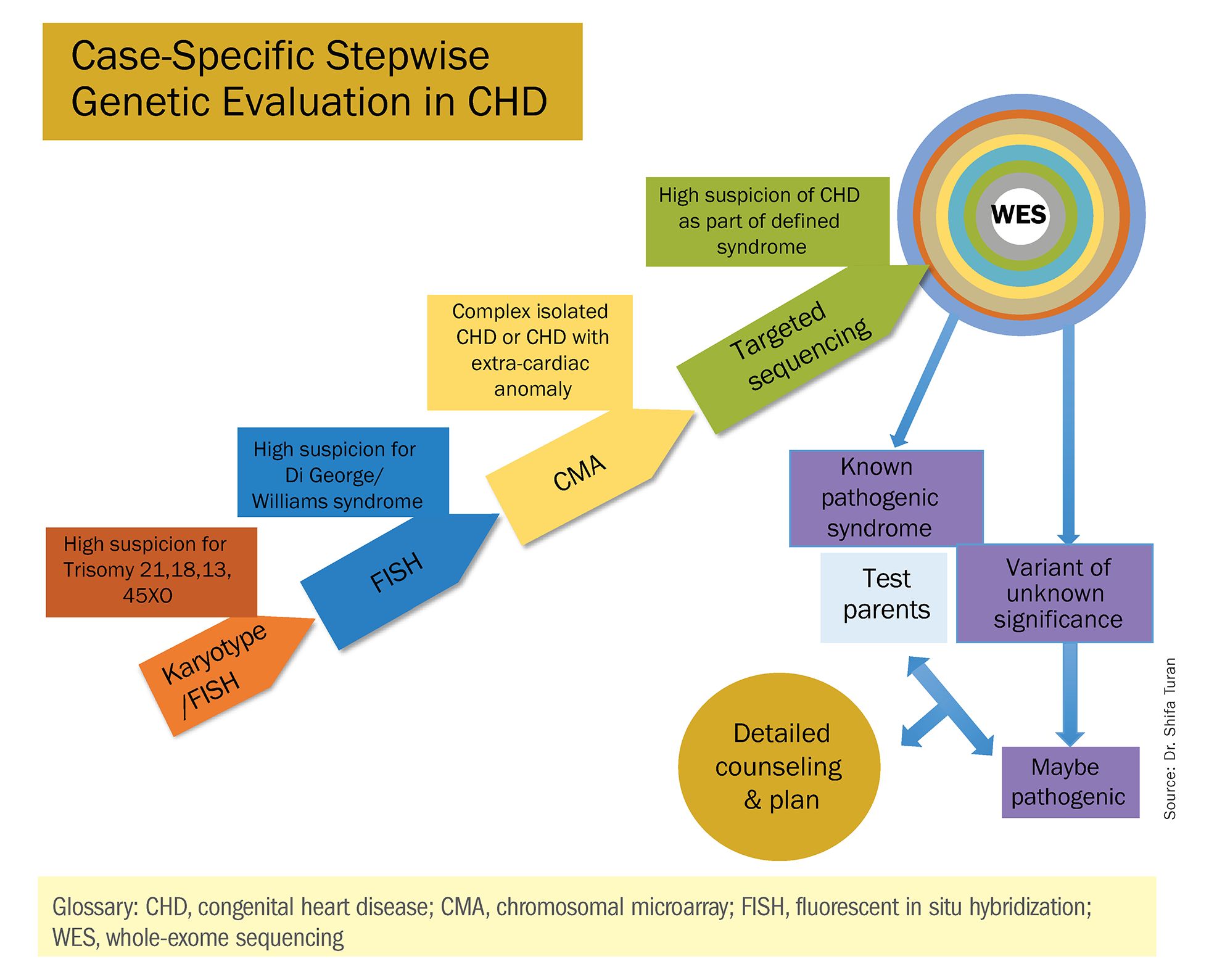
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.
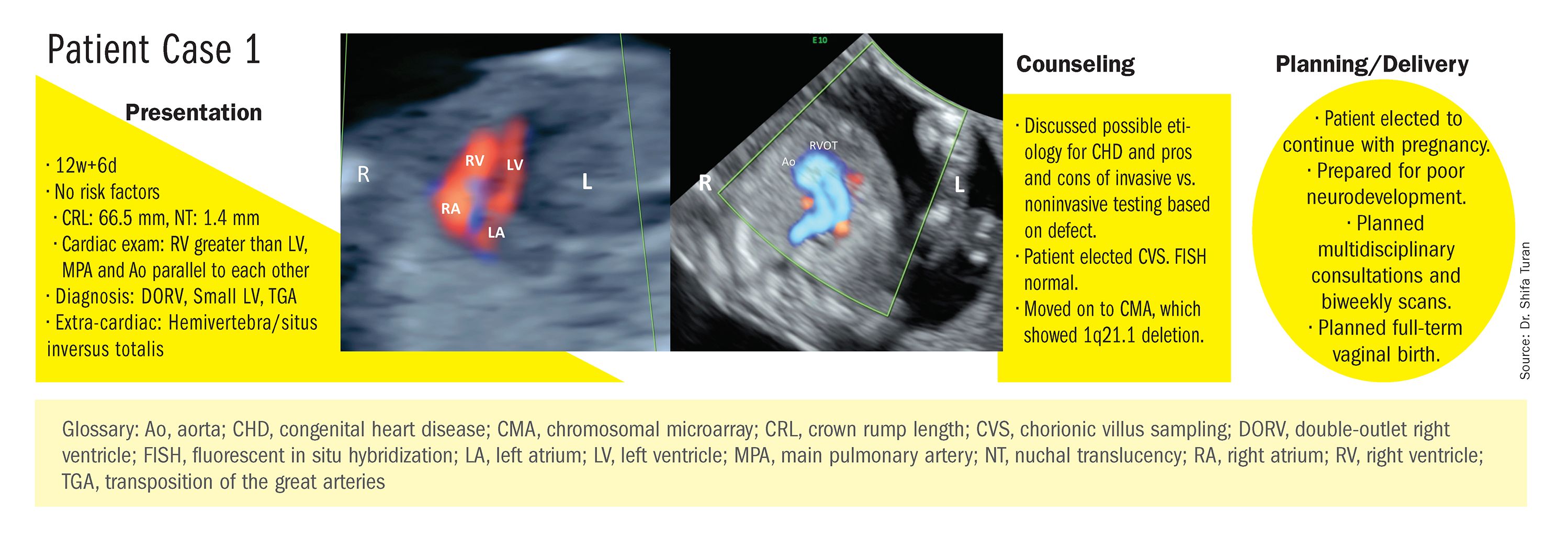
At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
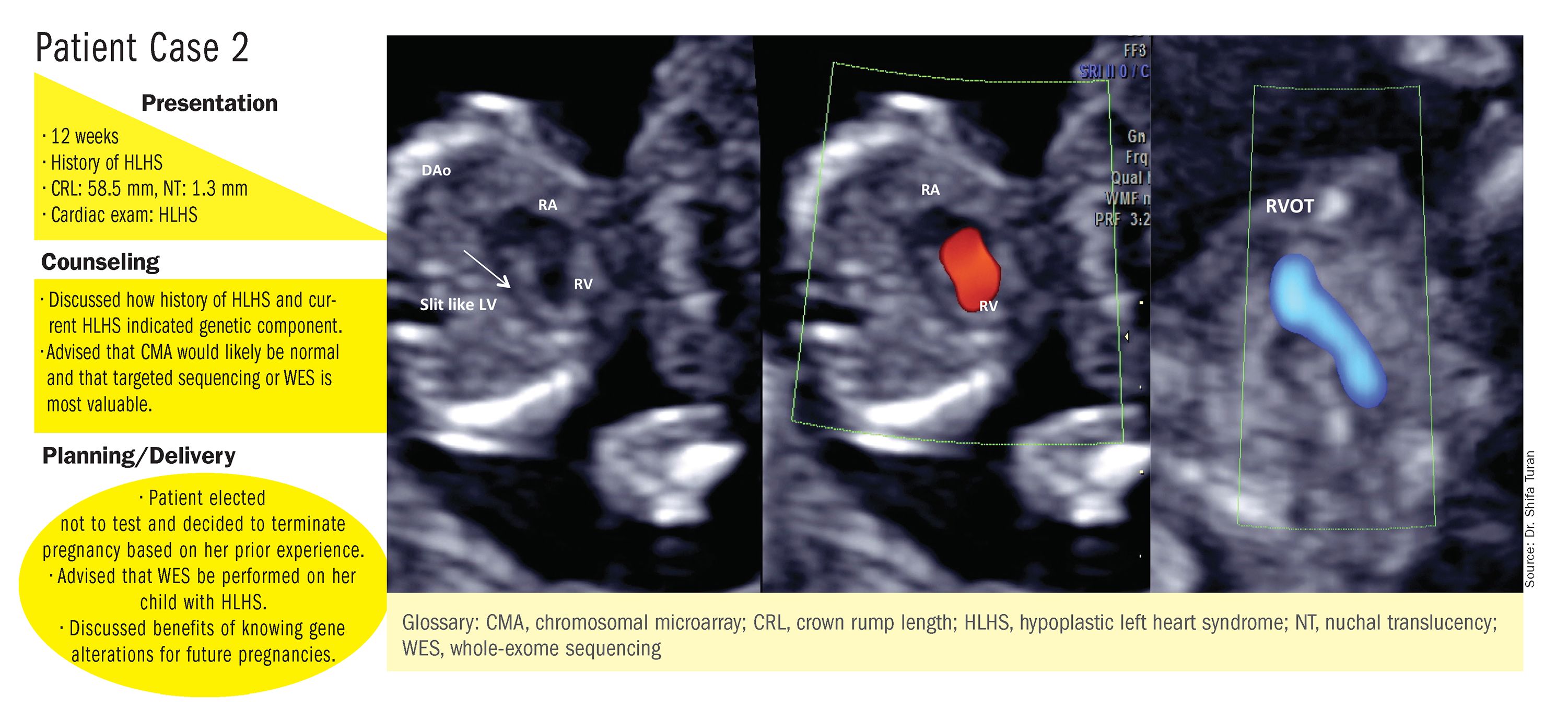
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.
These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.

At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.

These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.
Congenital heart defects (CHDs) are etiologically heterogeneous, but in recent years it has become clear that genetics plays a larger role in the development of CHDs than was previously thought. Research has been shifting from a focus on risk – estimating the magnitude of increased risk, for instance, based on maternal or familial risk factors – to a focus on the etiology of cardiac defects.
In practice, advances in genetic testing technologies have made the underlying causes of CHDs increasingly detectable. Chromosomal microarray analysis (CMA) – technology that detects significantly more and smaller changes in the amount of chromosomal material than traditional karyotype – has been proven to increase the diagnostic yield in cases of isolated CHDs and CHDs with extracardiac anomalies. Targeted next-generation sequencing also is now available as an additional approach in selective cases, and a clinically viable option for whole-exome sequencing is fast approaching.
For researchers, genetic evaluation carries the potential to unravel remaining mysteries about underlying causes of CHDs – to provide pathological insights and identify potential therapeutic targets. Currently, about 6 % of the total pie of presumed genetic determinants of CHDs is attributed to chromosomal anomalies, 10% to copy number variants, and 12% to single-gene defects. The remaining 72% of etiology, approximately, is undetermined.
As Helen Taussig, MD, (known as the founder of pediatric cardiology) once said, common cardiac malformations occurring in otherwise “normal” individuals “must be genetic in origin.”1 Greater use of genetic testing – and in particular, of whole-exome sequencing – will drive down this “undetermined” piece of the genetics pie.
For clinicians and patients, prenatal genetic evaluation can inform clinical management, guiding decisions on the mode, timing, and location of delivery. Genetic assessments help guide the neonatal health care team in taking optimal care of the infant, and the surgeon in preparing for neonatal surgeries and postsurgical complications.
In a recent analysis of the Society of Thoracic Surgeons Congenital Heart Surgery Database, prenatal diagnosis was associated with a lower overall prevalence of major preoperative risk factors for cardiac surgery.2 Surgical outcomes themselves also have been shown to be better after the prenatal diagnosis of complex CHDs, mainly because of improvements in perioperative care.3
When genetic etiology is elucidated, the cardiologist also is better able to counsel patients about anticipated challenges – such as the propensity, with certain genetic variants of CHD, to develop neurodevelopmental delays or other cardiac complications – and to target patient follow-up. Patients also can make informed decisions about termination or continuation of a current pregnancy and about family planning in the future.
Fortunately, advances in genetics technology have paralleled technological advancements in ultrasound. As I discussed in part one of this two-part Master Class series, it is now possible to detect many major CHDs well before 16 weeks’ gestation. Checking the structure of the fetal heart at the first-trimester screening and sonography (11-14 weeks of gestation) offers the opportunity for early genetic assessment, counseling, and planning when anomalies are detected.
A personalized approach
There has been growing interest in recent years in CMA for the prenatal genetic workup of CHDs. Microarray targets chromosomal regions at a much higher resolution than traditional karyotype. Traditional karyotype assesses both changes in chromosome number as well as more subtle structural changes such as chromosomal deletions and duplications. CMA finds what traditional karyotype identifies, but in addition, it identifies much smaller, clinically relevant chromosomal deletions and duplications that are not detected by karyotype performed with or without fluorescence in-situ hybridization (FISH). FISH uses DNA probes that carry fluorescent tags to detect chromosomal DNA.

At our center, we studied the prenatal genetic test results of 145 fetuses diagnosed with CHDs. Each case involved FISH for aneuploidy/karyotype, followed by CMA in cases of a negative karyotype result. CMA increased the diagnostic yield in cases of CHD by 19.8% overall – 17.4% in cases of isolated CHD and 24.5% in cases of CHD plus extracardiac anomalies.4
Indeed, although a microarray costs more and takes an additional 2 weeks to run, CMA should be strongly considered as first-line testing for the prenatal genetic evaluation of fetuses with major structural cardiac abnormalities detected by ultrasound. However, there still are cases in which a karyotype might be sufficient. For instance, if I see that a fetus has an atrial-ventricular septal defect on a prenatal ultrasound, and there are markers for trisomy 21, 13, or 18, or Turner’s syndrome (45 XO), I usually recommend a karyotype or FISH rather than an initial CMA. If the karyotype is abnormal – which is likely in such a scenario – there isn’t a need for more extensive testing.
Similarly, when there is high suspicion for DiGeorge syndrome (the 22q11.2 deletion, which often includes cleft palate and aortic arch abnormalities), usually it is most appropriate to perform a FISH test.
CMA is the preferred first modality, however, when prenatal imaging suggests severe CHD – for instance, when there are signs of hypoplastic left heart syndrome or tetralogy of Fallot (a conotruncal defect) – or complex CHD with extracardiac anomalies. In these cases, there is a high likelihood of detecting a small deletion or duplication that would be missed with karyotype.
In the past decade, karyotype and CMA have become the major methods used in our practice. However, targeted next‐generation sequencing and whole‐exome sequencing may become more widely used because these technologies enable rapid analysis of a large number of gene sequences and facilitate discovery of novel causative genes in many genetic diseases that cause CHDs.
Currently, targeted next-generation sequencing has mainly been used in the postnatal setting, and there are limited data available on its prenatal use. Compared with whole-exome sequencing, which sequences all of the protein-coding regions of the genome, targeted next-generation sequencing panels select regions of genes that are known to be associated with diseases of interest.
For CHDs, some perinatal centers have begun using a customized gene panel that targets 77 CHD-associated genes. This particular panel has been shown to be useful in addition to current methods and is an effective tool for prenatal genetic diagnosis.5
Whole-exome sequencing is currently expensive and time consuming. While sometimes it is used in the postnatal context, it is not yet part of routine practice as a prenatal diagnostic tool. As technology advances this will change – early in the next decade, I believe. For now, whole-exome sequencing may be an option for some patients who want to know more when severe CHD is evident on ultrasound and there are negative results from CMA or targeted sequencing. We have diagnosed some rare genetic syndromes using whole-exome sequencing; these diagnoses helped us to better manage the pregnancies.

These choices are part of the case-specific, stepwise approach to genetic evaluation that we take in our fetal heart program. Our goal is to pursue information that will be accurate and valuable for the patient and clinicians, in the most cost-effective and timely manner.
Limitations of noninvasive screening
In our fetal heart program we see increasing numbers of referred patients who have chosen noninvasive cell-free fetal DNA screening (cfDNA) after a cardiac anomaly is detected on ultrasound examination, and who believe that their “low risk” results demonstrate very little or no risk of CHD. Many of these patients express a belief that noninvasive testing is highly sensitive and accurate for fetal anomalies, including CHDs, and are not easily convinced of the value of other genetic tests.
We recently conducted a retrospective chart analysis (unpublished) in which we found that 41% of cases of CHD with abnormal genetics results were not detectable by cfDNA screening.
In the case of atrial-ventricular septal defects and conotruncal abnormalities that often are more associated with common aneuploidies (trisomy 21, 18, 13, and 45 XO), a “high-risk” result from cfDNA screening may offer the family and cardiology/neonatal team some guidance, but a “low-risk” result does not eliminate the risk of a microarray abnormality and thus may provide false reassurance.
Other research has shown that noninvasive screening will miss up to 7.3% of karyotype abnormalities in pregnancies at high risk for common aneuploidies.6
While invasive testing poses a very small risk of miscarriage, it is hard without such testing to elucidate the potential genetic etiologies of CHDs and truly understand the problems. We must take time to thoughtfully counsel patients who decline invasive testing about the limitations of cfDNA screening for CHDs and other anomalies.
Dr. Turan is an associate professor of obstetrics, gynecology, and reproductive sciences, and director of the fetal heart program at the University of Maryland School of Medicine and director of the Fetal Heart Program at the University of Maryland Medical Center. Dr. Turan reported that she has no disclosures relevant to this Master Class. Email her at obnews@mdedge.com.
References
1. J Am Coll Cardiol. 1988 Oct;12(4):1079-86.
2. Pediatr Cardiol. 2019 Mar;40(3):489-96.
3. Ann Pediatr Cardiol. 2017 May-Aug;10(2):126-30.
4. Eur J Obstet Gynecol Reprod Biol 2018;221:172-76.
5. Ultrasound Obstet Gynecol. 2018 Aug;52(2):205-11.
6. PLoS One. 2016 Jan 15;11(1):e0146794.

