User login
Making sense of CYP2D6 and CYP1A2 genotype vs phenotype
The clinical response to the same dose of a drug may vary among individuals. Cytochrome P450 (CYP) 2D6 and 1A2 are enzymes that metabolize many psychotropic medications. Genetic variations in these enzymes may cause changes in their activity and result in differences in effectiveness and adverse effects. Although pharmacogenetic testing is available for CYP2D6 and CYP1A2, interpretation and clinical application of the results may not be straightforward.
Genetic variations in CYP450 enzymes determine enzymatic activity, which can have a large effect on drug levels, efficacy, and toxicity. However, there are many other important factors that clinicians should consider when trying to predict the effects of medications. While clinicians often focus on a patient’s genotype, this only provides information on a chromosomal level, and this information never changes. In contrast, a patient’s phenotype, or status of metabolism, is subject to change throughout the patient’s life.
Many circumstances influence phenotype, including the use of medications that induce or inhibit CYP450 enzymes, environmental factors, and comorbidities. Phenoconversion occurs when these factors result in a phenotype that is different from that predicted by genotype. Because of the possibility of phenoconversion, knowing a patient’s genotype may be of limited value in making clinical decisions. This article provides guidance on interpreting both the genotype and phenotype of CYP2D6 and CYP1A2. Case 1 and Case 2 illustrate these concepts.
CYP2D6
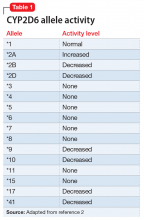
The enzyme activity of CYP2D6 varies among individuals and may include no activity, decreased activity, normal activity, or increased activity. After obtaining the genotype, the activity level of the CYP2D6 alleles may be determined. The frequency with which certain alleles occur varies with ancestry. More than 100 allelic variants and subvariants have been discovered, and new alleles are continuing to be discovered.1Table 12 lists some of the most common CYP2D6 alleles.
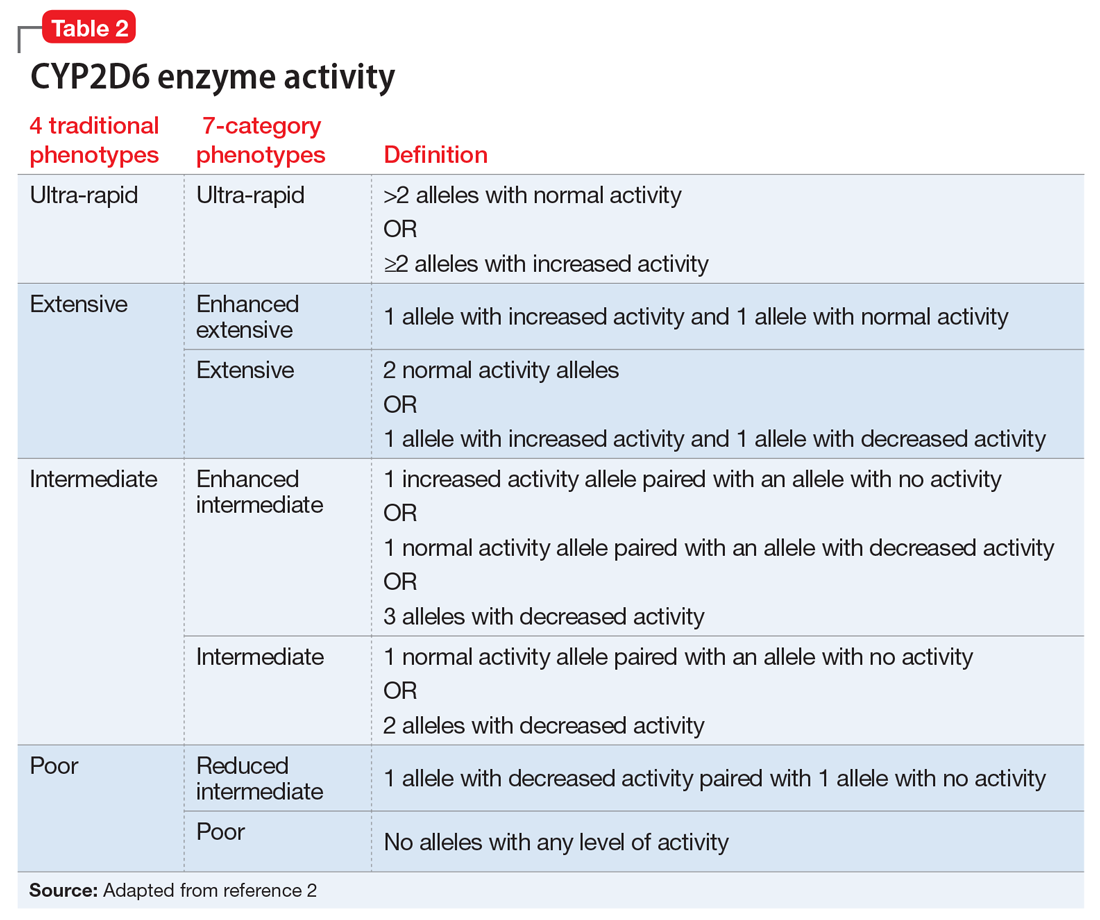
Based on the CYP2D6 enzyme activity determined from the alleles, 4 “traditional” phenotypes can be predicted from the genotype (Table 22). The 7-category phenotypes reported by some laboratory companies provide a more explicit method for reporting phenotypes.
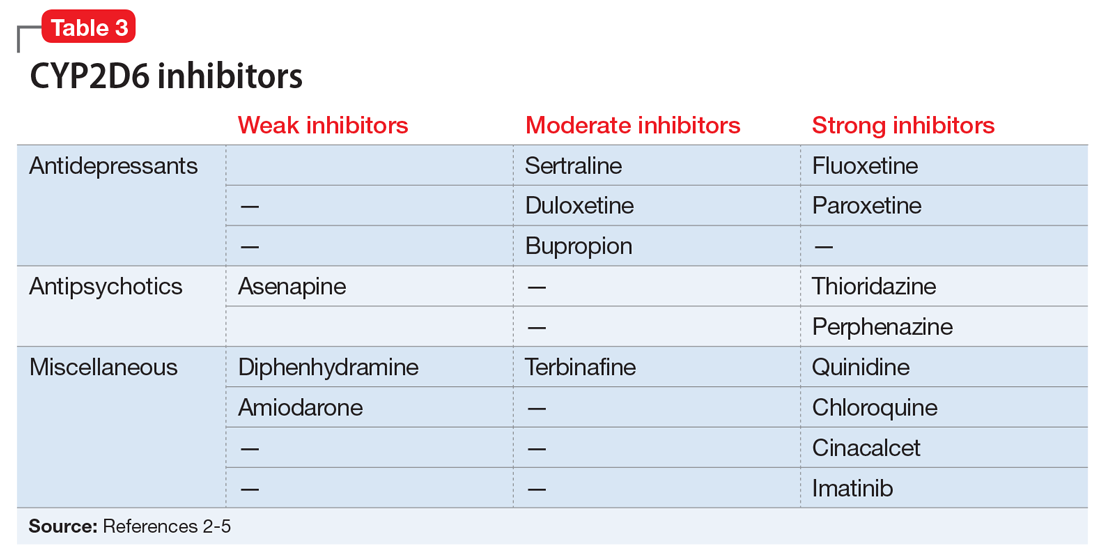
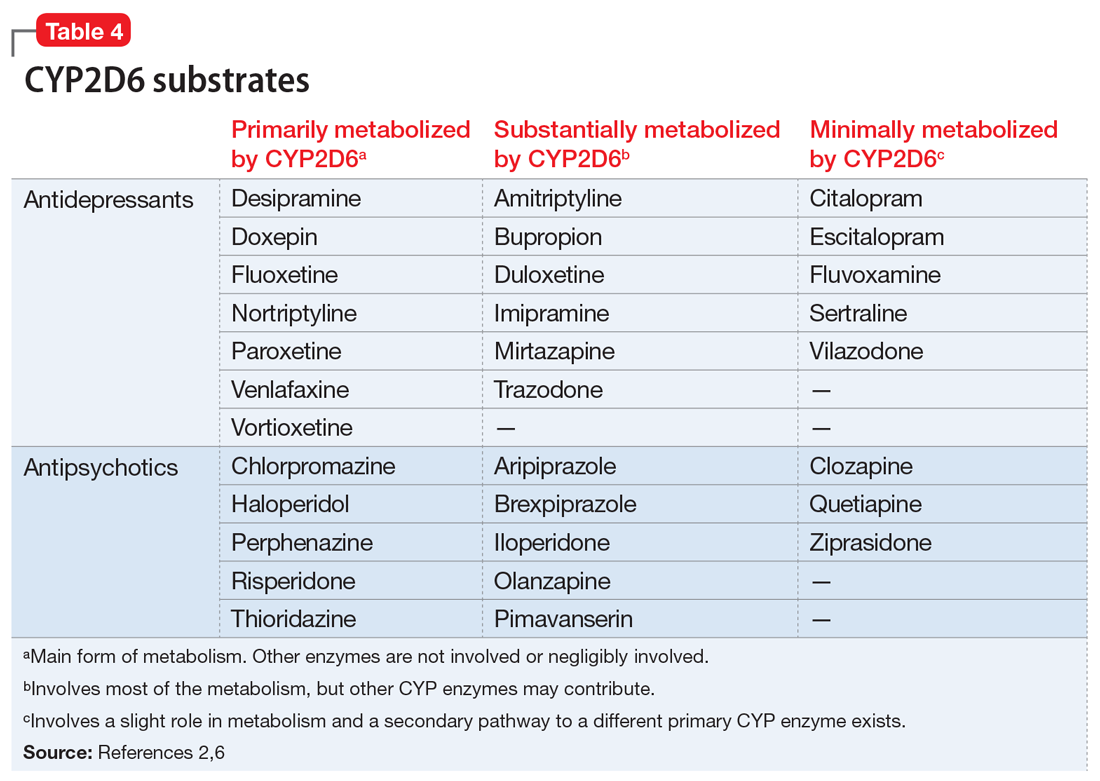
Evidence suggests that, unlike most other CYP450 enzymes, CYP2D6 is not very susceptible to enzyme induction.2 Thus, genetics, rather than drug therapy, accounts for most ultra-rapid CYP2D6 metabolizers. CYP2D6 can be inhibited by the use of medications (Table 32-5) and/or substrates (Table 42,6). Similar to inhibitors, substrates may be saturating high affinity-low capacity enzymes such as CYP2D6, resulting in phenoconversion to poor metabolizers. However, this is unlikely to be the case for substrates of low affinity-high capacity enzymes such as CYP3A4.7 Ultimately, substrates and/or inhibitors of CYP2D6 may result in a phenotype that does not correspond to genotype.
Phenoconversion
Genotyping may not reflect the true prevalence of the CYP2D6 poor metabolizer phenotype when using multiple medications that are substrates and/or inhibitors of CYP2D6.8 In the presence of strong CYP2D6 inhibitors, up to 80% of individuals with a non-poor metabolizer genotype are converted to a poor metabolizer phenotype.8 While the phenotype provides a clearer representation of metabolism status than genotype, this information may not always be available.
Continue to: Determining CYP2D6 phenotype
Determining CYP2D6 phenotype
Risperidone and venlafaxine levels are useful tools for predicting CYP2D6 phenotype.3,8 When a risperidone level is ordered, the results include a risperidone level and a 9-hydroxyrisperidone level. The active metabolite of risperidone is 9-hydroxyrisperidone (
- Ultra-rapid metabolizer: 0.03 (0.02 to 0.06)
- Extensive metabolizer: 0.08 (0.04 to 0.17)
- Intermediate metabolizer: 0.56 (0.30 to 1.0)
- Poor metabolizer: 2.5 (1.8 to 4.1).
Although a R-to-9-OHR concentration ratio >1 generally indicates a poor metabolizer, it could also indicate the presence of a powerful CYP2D6 inhibitor.9
When a venlafaxine level is ordered, the results include a venlafaxine level and an O-desmethylvenlafaxine level. O-desmethylvenlafaxine (
CYP1A2
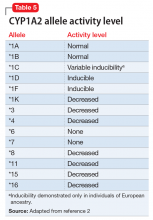
While the activity of CYP2D6 alleles is determined primarily by genetic factors and medications, the activity of CYP1A2 alleles is largely determined by environmental factors (diet, medications, disease) and genetic variability.2 Consequently, CYP1A2 genotyping may be less clinically useful than CYP2D6 genotyping. The CYP1A2 genotype–phenotype relationship incorporates the degree of allele activity (Table 52), and inducibility in the presence of environmental factors.
Continue to: CYP1A2 inhibiton
CYP1A2 inhibition
A variety of medications and environmental factors may inhibit CYP1A2.
Medications. Medications that may inhibit CYP1A2 include a
Caffeine. A significant increase in caffeine consumption can result in inhibition.3 Among non-tobacco smokers, an increase of 1 cup/d of coffee or 2 cans/d of caffeinated soda would be considered significant.3 However, tobacco smokers would require an increase of 3 cups/d of coffee or 6 cans/d of soda.
Diet. An increase in the daily dietary intake of certain vegetables for 6 days has been shown to result in inhibition.10 Apiaceous (Apiaceae or Umbelliferae) vegetables such as carrots (3/4 cup), celery (1/2 cup), dill (1 teaspoon), parsley (3 tablespoons), and parsnips (1¼ cup) can decrease CYP1A2 activity by approximately 13% to 25%. Allium (Liliaceae) vegetables, such as garlic, leeks, and onions, have no effect on CYP1A2 activity.
Infection. Pneumonia, upper respiratory infections with fever, pyelonephritis or appendicitis, or inflammation are suspected to decrease CYP1A2 activity.8
Continue to: CYP1A2 induction
CYP1A2 induction
A variety of medications and environmental factors may induce CYP1A2.
Medications. Certain medications may induce CYP1A2, including c
Cigarette smoking. A significant increase in smoking after 1 to 3 weeks may decrease drug levels, whereas a significant decrease in smoking after 1 to 3 weeks may result in elevated drug levels.3 Nicotine is not the causative agent of induction, but rather hydrocarbons found in cigarette smoke.11
Diet. An increase in daily dietary intake of certain vegetables for 6 days has been shown to result in induction.3 Brassica (Cruciferae) vegetables such as broccoli (2 cups), cauliflower (1 cup), cabbage (1 cup), and radish sprouts (1/2 cup) have been found to increase CYP1A2 activity by 18% to 37%.10 Grilled meat also plays a role in induction.10
Related Resource
- Ellingrod VL, Ward KM. Using pharmacogenetics guidelines when prescribing: What's available. Current Psychiatry. 2018;17(1):43-46.
Drug Brand Names
Amiodarone • Cordarone, Pacerone
Amitriptyline • Elavil, Endep
Aripiprazole • Abilify
Asenapine • Saphris
Atazanavir • Reyataz
Brexpiprazole • Rexulti
Bupropion • Wellbutrin, Zyban
Carbamazepine • Carbatrol, Tegretol
Chlorpromazine • Thorazine
Chloroquine • Aralen
Cinacalcet • Sensipar
Ciprofloxacin • Cipro
Citalopram • Celexa
Clozapine • Clozaril
Desipramine • Norpramin
Desvenlafaxine • Pristiq
Diphenhydramine • Benadryl
Doxepin • Silenor
Duloxetine • Cymbalta
Escitalopram • Lexapro
Ethinyl estradiol • Estinyl
Fluoxetine • Prozac
Fluvoxamine • Luvox
Haloperidol • Haldol
Iloperidone • Fanapt
Imatinib • Gleevec
Imipramine • Tofranil
Mirtazapine • Remeron
Nortriptyline • Pamelor
Olanzapine • Zyprexa
Paliperidone • Invega
Paroxetine • Paxil
Perphenazine • Trilafon
Phenytoin • Dilantin
Pimavanserin • Nuplazid
Primidone • Mysoline
Quetiapine • Seroquel
Quinidine • Cardioquin
Rifampin • Rifadin
Risperidone • Risperdal
Sertraline • Zoloft
Terbinafine • Lamisil
Thioridazine • Mellaril
Trazodone • Desyrel, Oleptro
Venlafaxine • Effexor
Vilazodone • Viibryd
Vortioxetine • Trintellix
Ziprasidone • Geodon
1. Pharmacogene Variation Consoritum. CYP2D6 allele nomenclature. https://www.pharmvar.org/gene/CYP2D6. Updated May 22, 2018. Accessed June 11, 2018.
2. Mrazek D. Psychiatric pharmacogenomics. New York, NY: Oxford University Press; 2010:33,42,44,45,85.
3. Spina E, de Leon J. Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna). 2015;122(1):5-28.
4. Adedoyin A, Frye RF, Mauro K, et al. Chloroquine modulation of specific metabolizing enzymes activities: investigation with selective five drug cocktail. Br J Clin Pharmacol. 1998;46(3):215-219.
5. Filppula AM, Laitila J, Neuvonen PJ, et al. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol. 2012;165(8):2787-2798.
6. U.S. National Library of Medicine. DailyMed. http://dailymed.nlm.nih.gov/dailymed/about.cfm. Accessed April 26, 2018.
7. Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014;21(8):879-885.
8. Preskorn SH, Kane CP, Lobello K, et al. Cytochrome P450 2D6 phenoconversion is common in patients being treated for depression: implications for personalized medicine. J Clin Psychiatry. 2013;74(6):614-621.
9. de Leon, J, Susce, MT, Johnson, M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr. 2009;14(1):19-34.
10. Lampe JW, King IB, Li S, et al. Brassica vegetables increase and apiaceous vegetables decrease cytochrome P450 1A2 activity in humans: changes in caffeine metabolite ratios in response to controlled vegetable diets. Carcinogenesis. 2000;21(6):1157-1162.
11. Zevin S, Benowitz NL. Drug interaction with tobacco smoking. An update. Clin Pharmacokinet. 1999;36(3):425-438.
The clinical response to the same dose of a drug may vary among individuals. Cytochrome P450 (CYP) 2D6 and 1A2 are enzymes that metabolize many psychotropic medications. Genetic variations in these enzymes may cause changes in their activity and result in differences in effectiveness and adverse effects. Although pharmacogenetic testing is available for CYP2D6 and CYP1A2, interpretation and clinical application of the results may not be straightforward.
Genetic variations in CYP450 enzymes determine enzymatic activity, which can have a large effect on drug levels, efficacy, and toxicity. However, there are many other important factors that clinicians should consider when trying to predict the effects of medications. While clinicians often focus on a patient’s genotype, this only provides information on a chromosomal level, and this information never changes. In contrast, a patient’s phenotype, or status of metabolism, is subject to change throughout the patient’s life.
Many circumstances influence phenotype, including the use of medications that induce or inhibit CYP450 enzymes, environmental factors, and comorbidities. Phenoconversion occurs when these factors result in a phenotype that is different from that predicted by genotype. Because of the possibility of phenoconversion, knowing a patient’s genotype may be of limited value in making clinical decisions. This article provides guidance on interpreting both the genotype and phenotype of CYP2D6 and CYP1A2. Case 1 and Case 2 illustrate these concepts.
CYP2D6
The enzyme activity of CYP2D6 varies among individuals and may include no activity, decreased activity, normal activity, or increased activity. After obtaining the genotype, the activity level of the CYP2D6 alleles may be determined. The frequency with which certain alleles occur varies with ancestry. More than 100 allelic variants and subvariants have been discovered, and new alleles are continuing to be discovered.1Table 12 lists some of the most common CYP2D6 alleles.
Based on the CYP2D6 enzyme activity determined from the alleles, 4 “traditional” phenotypes can be predicted from the genotype (Table 22). The 7-category phenotypes reported by some laboratory companies provide a more explicit method for reporting phenotypes.
Evidence suggests that, unlike most other CYP450 enzymes, CYP2D6 is not very susceptible to enzyme induction.2 Thus, genetics, rather than drug therapy, accounts for most ultra-rapid CYP2D6 metabolizers. CYP2D6 can be inhibited by the use of medications (Table 32-5) and/or substrates (Table 42,6). Similar to inhibitors, substrates may be saturating high affinity-low capacity enzymes such as CYP2D6, resulting in phenoconversion to poor metabolizers. However, this is unlikely to be the case for substrates of low affinity-high capacity enzymes such as CYP3A4.7 Ultimately, substrates and/or inhibitors of CYP2D6 may result in a phenotype that does not correspond to genotype.
Phenoconversion
Genotyping may not reflect the true prevalence of the CYP2D6 poor metabolizer phenotype when using multiple medications that are substrates and/or inhibitors of CYP2D6.8 In the presence of strong CYP2D6 inhibitors, up to 80% of individuals with a non-poor metabolizer genotype are converted to a poor metabolizer phenotype.8 While the phenotype provides a clearer representation of metabolism status than genotype, this information may not always be available.
Continue to: Determining CYP2D6 phenotype
Determining CYP2D6 phenotype
Risperidone and venlafaxine levels are useful tools for predicting CYP2D6 phenotype.3,8 When a risperidone level is ordered, the results include a risperidone level and a 9-hydroxyrisperidone level. The active metabolite of risperidone is 9-hydroxyrisperidone (
- Ultra-rapid metabolizer: 0.03 (0.02 to 0.06)
- Extensive metabolizer: 0.08 (0.04 to 0.17)
- Intermediate metabolizer: 0.56 (0.30 to 1.0)
- Poor metabolizer: 2.5 (1.8 to 4.1).
Although a R-to-9-OHR concentration ratio >1 generally indicates a poor metabolizer, it could also indicate the presence of a powerful CYP2D6 inhibitor.9
When a venlafaxine level is ordered, the results include a venlafaxine level and an O-desmethylvenlafaxine level. O-desmethylvenlafaxine (
CYP1A2
While the activity of CYP2D6 alleles is determined primarily by genetic factors and medications, the activity of CYP1A2 alleles is largely determined by environmental factors (diet, medications, disease) and genetic variability.2 Consequently, CYP1A2 genotyping may be less clinically useful than CYP2D6 genotyping. The CYP1A2 genotype–phenotype relationship incorporates the degree of allele activity (Table 52), and inducibility in the presence of environmental factors.
Continue to: CYP1A2 inhibiton
CYP1A2 inhibition
A variety of medications and environmental factors may inhibit CYP1A2.
Medications. Medications that may inhibit CYP1A2 include a
Caffeine. A significant increase in caffeine consumption can result in inhibition.3 Among non-tobacco smokers, an increase of 1 cup/d of coffee or 2 cans/d of caffeinated soda would be considered significant.3 However, tobacco smokers would require an increase of 3 cups/d of coffee or 6 cans/d of soda.
Diet. An increase in the daily dietary intake of certain vegetables for 6 days has been shown to result in inhibition.10 Apiaceous (Apiaceae or Umbelliferae) vegetables such as carrots (3/4 cup), celery (1/2 cup), dill (1 teaspoon), parsley (3 tablespoons), and parsnips (1¼ cup) can decrease CYP1A2 activity by approximately 13% to 25%. Allium (Liliaceae) vegetables, such as garlic, leeks, and onions, have no effect on CYP1A2 activity.
Infection. Pneumonia, upper respiratory infections with fever, pyelonephritis or appendicitis, or inflammation are suspected to decrease CYP1A2 activity.8
Continue to: CYP1A2 induction
CYP1A2 induction
A variety of medications and environmental factors may induce CYP1A2.
Medications. Certain medications may induce CYP1A2, including c
Cigarette smoking. A significant increase in smoking after 1 to 3 weeks may decrease drug levels, whereas a significant decrease in smoking after 1 to 3 weeks may result in elevated drug levels.3 Nicotine is not the causative agent of induction, but rather hydrocarbons found in cigarette smoke.11
Diet. An increase in daily dietary intake of certain vegetables for 6 days has been shown to result in induction.3 Brassica (Cruciferae) vegetables such as broccoli (2 cups), cauliflower (1 cup), cabbage (1 cup), and radish sprouts (1/2 cup) have been found to increase CYP1A2 activity by 18% to 37%.10 Grilled meat also plays a role in induction.10
Related Resource
- Ellingrod VL, Ward KM. Using pharmacogenetics guidelines when prescribing: What's available. Current Psychiatry. 2018;17(1):43-46.
Drug Brand Names
Amiodarone • Cordarone, Pacerone
Amitriptyline • Elavil, Endep
Aripiprazole • Abilify
Asenapine • Saphris
Atazanavir • Reyataz
Brexpiprazole • Rexulti
Bupropion • Wellbutrin, Zyban
Carbamazepine • Carbatrol, Tegretol
Chlorpromazine • Thorazine
Chloroquine • Aralen
Cinacalcet • Sensipar
Ciprofloxacin • Cipro
Citalopram • Celexa
Clozapine • Clozaril
Desipramine • Norpramin
Desvenlafaxine • Pristiq
Diphenhydramine • Benadryl
Doxepin • Silenor
Duloxetine • Cymbalta
Escitalopram • Lexapro
Ethinyl estradiol • Estinyl
Fluoxetine • Prozac
Fluvoxamine • Luvox
Haloperidol • Haldol
Iloperidone • Fanapt
Imatinib • Gleevec
Imipramine • Tofranil
Mirtazapine • Remeron
Nortriptyline • Pamelor
Olanzapine • Zyprexa
Paliperidone • Invega
Paroxetine • Paxil
Perphenazine • Trilafon
Phenytoin • Dilantin
Pimavanserin • Nuplazid
Primidone • Mysoline
Quetiapine • Seroquel
Quinidine • Cardioquin
Rifampin • Rifadin
Risperidone • Risperdal
Sertraline • Zoloft
Terbinafine • Lamisil
Thioridazine • Mellaril
Trazodone • Desyrel, Oleptro
Venlafaxine • Effexor
Vilazodone • Viibryd
Vortioxetine • Trintellix
Ziprasidone • Geodon
The clinical response to the same dose of a drug may vary among individuals. Cytochrome P450 (CYP) 2D6 and 1A2 are enzymes that metabolize many psychotropic medications. Genetic variations in these enzymes may cause changes in their activity and result in differences in effectiveness and adverse effects. Although pharmacogenetic testing is available for CYP2D6 and CYP1A2, interpretation and clinical application of the results may not be straightforward.
Genetic variations in CYP450 enzymes determine enzymatic activity, which can have a large effect on drug levels, efficacy, and toxicity. However, there are many other important factors that clinicians should consider when trying to predict the effects of medications. While clinicians often focus on a patient’s genotype, this only provides information on a chromosomal level, and this information never changes. In contrast, a patient’s phenotype, or status of metabolism, is subject to change throughout the patient’s life.
Many circumstances influence phenotype, including the use of medications that induce or inhibit CYP450 enzymes, environmental factors, and comorbidities. Phenoconversion occurs when these factors result in a phenotype that is different from that predicted by genotype. Because of the possibility of phenoconversion, knowing a patient’s genotype may be of limited value in making clinical decisions. This article provides guidance on interpreting both the genotype and phenotype of CYP2D6 and CYP1A2. Case 1 and Case 2 illustrate these concepts.
CYP2D6
The enzyme activity of CYP2D6 varies among individuals and may include no activity, decreased activity, normal activity, or increased activity. After obtaining the genotype, the activity level of the CYP2D6 alleles may be determined. The frequency with which certain alleles occur varies with ancestry. More than 100 allelic variants and subvariants have been discovered, and new alleles are continuing to be discovered.1Table 12 lists some of the most common CYP2D6 alleles.
Based on the CYP2D6 enzyme activity determined from the alleles, 4 “traditional” phenotypes can be predicted from the genotype (Table 22). The 7-category phenotypes reported by some laboratory companies provide a more explicit method for reporting phenotypes.
Evidence suggests that, unlike most other CYP450 enzymes, CYP2D6 is not very susceptible to enzyme induction.2 Thus, genetics, rather than drug therapy, accounts for most ultra-rapid CYP2D6 metabolizers. CYP2D6 can be inhibited by the use of medications (Table 32-5) and/or substrates (Table 42,6). Similar to inhibitors, substrates may be saturating high affinity-low capacity enzymes such as CYP2D6, resulting in phenoconversion to poor metabolizers. However, this is unlikely to be the case for substrates of low affinity-high capacity enzymes such as CYP3A4.7 Ultimately, substrates and/or inhibitors of CYP2D6 may result in a phenotype that does not correspond to genotype.
Phenoconversion
Genotyping may not reflect the true prevalence of the CYP2D6 poor metabolizer phenotype when using multiple medications that are substrates and/or inhibitors of CYP2D6.8 In the presence of strong CYP2D6 inhibitors, up to 80% of individuals with a non-poor metabolizer genotype are converted to a poor metabolizer phenotype.8 While the phenotype provides a clearer representation of metabolism status than genotype, this information may not always be available.
Continue to: Determining CYP2D6 phenotype
Determining CYP2D6 phenotype
Risperidone and venlafaxine levels are useful tools for predicting CYP2D6 phenotype.3,8 When a risperidone level is ordered, the results include a risperidone level and a 9-hydroxyrisperidone level. The active metabolite of risperidone is 9-hydroxyrisperidone (
- Ultra-rapid metabolizer: 0.03 (0.02 to 0.06)
- Extensive metabolizer: 0.08 (0.04 to 0.17)
- Intermediate metabolizer: 0.56 (0.30 to 1.0)
- Poor metabolizer: 2.5 (1.8 to 4.1).
Although a R-to-9-OHR concentration ratio >1 generally indicates a poor metabolizer, it could also indicate the presence of a powerful CYP2D6 inhibitor.9
When a venlafaxine level is ordered, the results include a venlafaxine level and an O-desmethylvenlafaxine level. O-desmethylvenlafaxine (
CYP1A2
While the activity of CYP2D6 alleles is determined primarily by genetic factors and medications, the activity of CYP1A2 alleles is largely determined by environmental factors (diet, medications, disease) and genetic variability.2 Consequently, CYP1A2 genotyping may be less clinically useful than CYP2D6 genotyping. The CYP1A2 genotype–phenotype relationship incorporates the degree of allele activity (Table 52), and inducibility in the presence of environmental factors.
Continue to: CYP1A2 inhibiton
CYP1A2 inhibition
A variety of medications and environmental factors may inhibit CYP1A2.
Medications. Medications that may inhibit CYP1A2 include a
Caffeine. A significant increase in caffeine consumption can result in inhibition.3 Among non-tobacco smokers, an increase of 1 cup/d of coffee or 2 cans/d of caffeinated soda would be considered significant.3 However, tobacco smokers would require an increase of 3 cups/d of coffee or 6 cans/d of soda.
Diet. An increase in the daily dietary intake of certain vegetables for 6 days has been shown to result in inhibition.10 Apiaceous (Apiaceae or Umbelliferae) vegetables such as carrots (3/4 cup), celery (1/2 cup), dill (1 teaspoon), parsley (3 tablespoons), and parsnips (1¼ cup) can decrease CYP1A2 activity by approximately 13% to 25%. Allium (Liliaceae) vegetables, such as garlic, leeks, and onions, have no effect on CYP1A2 activity.
Infection. Pneumonia, upper respiratory infections with fever, pyelonephritis or appendicitis, or inflammation are suspected to decrease CYP1A2 activity.8
Continue to: CYP1A2 induction
CYP1A2 induction
A variety of medications and environmental factors may induce CYP1A2.
Medications. Certain medications may induce CYP1A2, including c
Cigarette smoking. A significant increase in smoking after 1 to 3 weeks may decrease drug levels, whereas a significant decrease in smoking after 1 to 3 weeks may result in elevated drug levels.3 Nicotine is not the causative agent of induction, but rather hydrocarbons found in cigarette smoke.11
Diet. An increase in daily dietary intake of certain vegetables for 6 days has been shown to result in induction.3 Brassica (Cruciferae) vegetables such as broccoli (2 cups), cauliflower (1 cup), cabbage (1 cup), and radish sprouts (1/2 cup) have been found to increase CYP1A2 activity by 18% to 37%.10 Grilled meat also plays a role in induction.10
Related Resource
- Ellingrod VL, Ward KM. Using pharmacogenetics guidelines when prescribing: What's available. Current Psychiatry. 2018;17(1):43-46.
Drug Brand Names
Amiodarone • Cordarone, Pacerone
Amitriptyline • Elavil, Endep
Aripiprazole • Abilify
Asenapine • Saphris
Atazanavir • Reyataz
Brexpiprazole • Rexulti
Bupropion • Wellbutrin, Zyban
Carbamazepine • Carbatrol, Tegretol
Chlorpromazine • Thorazine
Chloroquine • Aralen
Cinacalcet • Sensipar
Ciprofloxacin • Cipro
Citalopram • Celexa
Clozapine • Clozaril
Desipramine • Norpramin
Desvenlafaxine • Pristiq
Diphenhydramine • Benadryl
Doxepin • Silenor
Duloxetine • Cymbalta
Escitalopram • Lexapro
Ethinyl estradiol • Estinyl
Fluoxetine • Prozac
Fluvoxamine • Luvox
Haloperidol • Haldol
Iloperidone • Fanapt
Imatinib • Gleevec
Imipramine • Tofranil
Mirtazapine • Remeron
Nortriptyline • Pamelor
Olanzapine • Zyprexa
Paliperidone • Invega
Paroxetine • Paxil
Perphenazine • Trilafon
Phenytoin • Dilantin
Pimavanserin • Nuplazid
Primidone • Mysoline
Quetiapine • Seroquel
Quinidine • Cardioquin
Rifampin • Rifadin
Risperidone • Risperdal
Sertraline • Zoloft
Terbinafine • Lamisil
Thioridazine • Mellaril
Trazodone • Desyrel, Oleptro
Venlafaxine • Effexor
Vilazodone • Viibryd
Vortioxetine • Trintellix
Ziprasidone • Geodon
1. Pharmacogene Variation Consoritum. CYP2D6 allele nomenclature. https://www.pharmvar.org/gene/CYP2D6. Updated May 22, 2018. Accessed June 11, 2018.
2. Mrazek D. Psychiatric pharmacogenomics. New York, NY: Oxford University Press; 2010:33,42,44,45,85.
3. Spina E, de Leon J. Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna). 2015;122(1):5-28.
4. Adedoyin A, Frye RF, Mauro K, et al. Chloroquine modulation of specific metabolizing enzymes activities: investigation with selective five drug cocktail. Br J Clin Pharmacol. 1998;46(3):215-219.
5. Filppula AM, Laitila J, Neuvonen PJ, et al. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol. 2012;165(8):2787-2798.
6. U.S. National Library of Medicine. DailyMed. http://dailymed.nlm.nih.gov/dailymed/about.cfm. Accessed April 26, 2018.
7. Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014;21(8):879-885.
8. Preskorn SH, Kane CP, Lobello K, et al. Cytochrome P450 2D6 phenoconversion is common in patients being treated for depression: implications for personalized medicine. J Clin Psychiatry. 2013;74(6):614-621.
9. de Leon, J, Susce, MT, Johnson, M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr. 2009;14(1):19-34.
10. Lampe JW, King IB, Li S, et al. Brassica vegetables increase and apiaceous vegetables decrease cytochrome P450 1A2 activity in humans: changes in caffeine metabolite ratios in response to controlled vegetable diets. Carcinogenesis. 2000;21(6):1157-1162.
11. Zevin S, Benowitz NL. Drug interaction with tobacco smoking. An update. Clin Pharmacokinet. 1999;36(3):425-438.
1. Pharmacogene Variation Consoritum. CYP2D6 allele nomenclature. https://www.pharmvar.org/gene/CYP2D6. Updated May 22, 2018. Accessed June 11, 2018.
2. Mrazek D. Psychiatric pharmacogenomics. New York, NY: Oxford University Press; 2010:33,42,44,45,85.
3. Spina E, de Leon J. Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna). 2015;122(1):5-28.
4. Adedoyin A, Frye RF, Mauro K, et al. Chloroquine modulation of specific metabolizing enzymes activities: investigation with selective five drug cocktail. Br J Clin Pharmacol. 1998;46(3):215-219.
5. Filppula AM, Laitila J, Neuvonen PJ, et al. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol. 2012;165(8):2787-2798.
6. U.S. National Library of Medicine. DailyMed. http://dailymed.nlm.nih.gov/dailymed/about.cfm. Accessed April 26, 2018.
7. Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014;21(8):879-885.
8. Preskorn SH, Kane CP, Lobello K, et al. Cytochrome P450 2D6 phenoconversion is common in patients being treated for depression: implications for personalized medicine. J Clin Psychiatry. 2013;74(6):614-621.
9. de Leon, J, Susce, MT, Johnson, M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr. 2009;14(1):19-34.
10. Lampe JW, King IB, Li S, et al. Brassica vegetables increase and apiaceous vegetables decrease cytochrome P450 1A2 activity in humans: changes in caffeine metabolite ratios in response to controlled vegetable diets. Carcinogenesis. 2000;21(6):1157-1162.
11. Zevin S, Benowitz NL. Drug interaction with tobacco smoking. An update. Clin Pharmacokinet. 1999;36(3):425-438.