User login
Whole exome sequencing’s role in diagnosing genetic causes of FGR with and without associated anomalies
Mone F, Mellis R, Gabriel H, et al. Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am J Obstet Gynecol. Published online October 7, 2022. doi:10.1016/j.ajog.2022.09.045
Multiple factors can play a role in FGR, including inherent maternal, placental, or fetal factors; the environment; and/or nutrition. However, prenatal diagnosis is an important consideration when exploring the underlying etiology for a growth-restricted fetus, especially in severe or early-onset cases. Many genetic conditions do not result in structural anomalies but can disrupt overall growth. Additionally, phenotyping in the prenatal period is limited and can miss more subtle physical differences that could point to a genetic cause.
When compared with karyotype, chromosomal microarray (CMA) has been shown to increase the diagnostic yield in cases of isolated early FGR by 5%,1,2 and the incidence of chromosomal abnormalities has been reported to be as high as 19% in this population. Let’s explore the data on exome sequencing for prenatal diagnosis in cases of isolated FGR.
Meta-analysis details
In this meta-analysis, the authors reviewed 19 cohort studies or case series that investigated the yield of prenatal sequencing in fetuses with intrauterine growth restriction (IUGR) or short long bones, both in association with and without additional anomalies. All cases had nondiagnostic cytogenetic results. Fetal DNA in most cases was obtained through amniocentesis. Variants classified as likely pathogenic and pathogenic were considered diagnostic. The authors then calculated the incremental yield of prenatal sequencing over cytogenetic studies as a pooled value, comparing the following groups:
- isolated FGR
- growth restriction with associated anomalies
- isolated short long bones
- short long bones with additional skeletal features.
Study outcomes
The total number of cases were as follows: isolated IUGR (n = 71), IUGR associated with additional anomalies (n = 45), isolated short long bones (n = 84), and short long bones associated with additional skeletal findings (n = 252). Causative pathogenic or likely pathogenic variants were identified in 224 (50%) cases. Apparent incremental yields with prenatal sequencing were as follows for the 4 groups (as illustrated in the FIGURE):
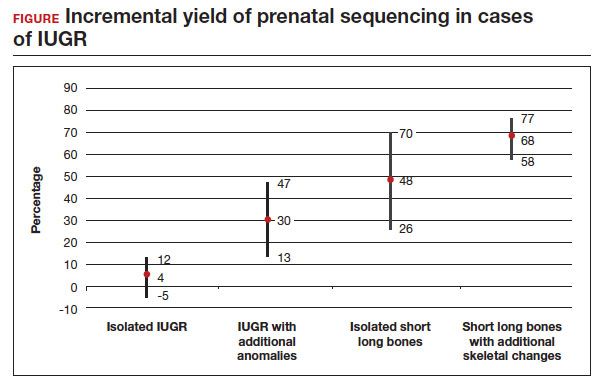
- 4% in isolated IUGR (95% confidence interval [CI], -5%–12%)
- 30% in IUGR with additional anomalies (95% CI, 13%–47%)
- 48% in isolated short long bones (95% CI, 26%–70%)
- 68% in short long bones with additional skeletal changes (95% CI, 58%–77%).
Overall, the authors concluded that prenatal sequencing does not improve prenatal diagnosis in cases of isolated IUGR. The majority of these cases were thought to be related to placental insufficiency.
Strengths and limitations
The main limitation of this study with regard to our discussion is the small study populationof isolated growth restriction. The authors indicate that the number of cases of isolated IUGR were too small to draw firm conclusions. Another limitation was the heterogeneity of the isolated FGR population, which was not limited to severe or early-onset cases. However, the authors did demonstrate that growth restriction in association with fetal anomalies has very high genetic yield rates with prenatal sequencing.
Not surprisingly, there is a high yield of diagnosing genetic conditions in pregnancies complicated by isolated or nonisolated short long bones or in cases of growth restriction with multisystem abnormalities. Based on the results of this study, the authors advise against sending for exome sequencing in cases of isolated growth restriction with coexisting evidence of placental insufficiency.
Continue to: Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?...
Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?
Zhou H, Fu F, Wang Y, et al. Genetic causes of isolated and severe fetal growth restriction in normal chromosomal microarray analysis. Int J Gynaecol Obstet. Published online December 10, 2022. doi:10.1002/ijgo.14620
Severe FGR is diagnosed based on an estimated fetal weight (EFW) or abdominal circumference (AC) below the third percentile. As we discussed in the above study by Mone and colleagues, it does not appear that prenatal sequencing significantly improves the diagnostic yield in all isolated FGR cases. However, this has not been previously explored in isolated severe FGR or cases of early-onset FGR (<32 weeks’ gestation). We know that several monogenic conditions are associated with severe and early-onset isolated fetal growth impairment, including but not limited to Cornelia de Lange syndrome, Smith-Lemli-Opitz syndrome, and Meier-Gorlin syndrome. Often, these syndromes can present in the prenatal period without other phenotypic findings. Therefore, this study explored the possibility that prenatal sequencing plays an important role for severe cases of FGR with nondiagnostic CMA and/or karyotype.
Retrospective study details
Zhou and colleagues retrospectively analyzed 51 cases of severe (EFW or AC below the third percentile) isolated FGR with negative CMA who underwent trio whole exome sequencing, which includes submitting fetal cells as well as both parental samples for testing. Patients with abnormal toxoplasmosis, rubella, cytomegalovirus, and herpes simplex virus (TORCH) tests; structural anomalies; and multiple gestation were excluded from the analysis. As in the study by Mone et al, variants classified as likely pathogenic and pathogenic were categorized as diagnostic.
Results
Eight of 51 cases (15.7%) with severe isolated FGR had diagnostic findings on trio whole exome sequencing as shown in the TABLE. Another 8 cases (15.7%) were found to have variants of unknown significance, of which 2 were later determined to be novel pathogenic variants. Genetic conditions uncovered in this cohort include Cornelia de Lange syndrome, pyruvate dehydrogenase deficiency, Dent disease, trichohepaticenteric syndrome, achondroplasia, osteogenesis imperfecta, Pendred syndrome, and both autosomal dominant type 3A and autosomal recessive type 1A deafness. All 10 cases with diagnostic whole exome sequencing or identified novel pathogenic variants were affected by early-onset FGR (<32 weeks’ gestation). Of these 10 cases, 7 patients underwent pregnancy termination.
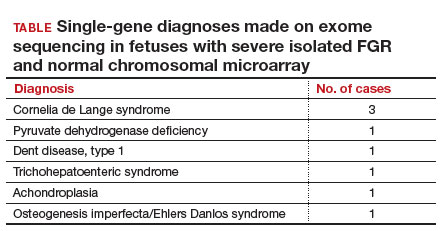
To summarize, a total of 10 cases (19.6%) of severe isolated early-onset FGR with negative cytogenetic studies were subsequently diagnosed with an underlying genetic condition using prenatal trio whole exome sequencing.
Strengths and limitations
This study is retrospective and has a small sample size (n = 51) that was mostly limited to early-onset isolated severe FGR. However, the diagnostic yield (19.6%) of whole exome sequencing after negative CMA testing was noteworthy and shows that monogenic conditions are an important consideration in the evaluation of severe early-onset FGR, even in the absence of structural abnormalities.
As indications for exome sequencing during pregnancy continue to evolve, severe isolated FGR is emerging as a high-yield condition in which a subset of patients may benefit from the described testing strategy. We learned from our look at the prior study (Mone et al) that unselected isolated growth restriction with evident placental insufficiency may not benefit from exome sequencing, but this study differs in its selection of early-onset, severe cases—defined by diagnosis before 32 weeks’ gestation and an EFW or AC below the third percentile. Almost 20% of cases who met the aforementioned criteria received a genetic diagnosis from exome sequencing. We should remember to offer genetic counseling and diagnostic testing to our patients with severe growth restriction, even in the absence of additional structural anomalies.
Could epigenetic mechanisms of placental dysregulation explain low birthweight and future cardiometabolic disease?
Tekola-Ayele F, Zeng X, Chatterjee S, et al. Placental multi-omics integration identifies candidate functional genes for birthweight. Nat Commun. 2022;13:2384.
FGR has been linked to greater mortality in childhood and increased risk for cardiometabolic disease in adulthood. While genomewide associations studies (GWAS) have defined areas of interest linking genetic variants to low birthweight, their relationship to epigenetic changes in the placenta as well as biologic and functional mechanisms are not yet well understood.
Multiomics used to identify candidate functional genes for birthweight
This study analyzed the methylation and gene expression patterns of 291 placental samples, integrating findings into pathways of previously defined GWAS variants. Patient samples were obtained from participants in the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Fetal Growth Studies–Singleton cohort. The cohort is ethnically diverse, with 97 Hispanic, 74 White, 71 Black, and 49 Asian participants. Of 286 single nucleotide polymorphisms (SNPs) found to be associated with birthweight, 273 were analyzed as part of the authors’ data set. These were found to have 7,901 unique protein-coding mRNAs (expression quantitative trait loci [eQTL]) and more than 100,000 nearby (within 1 Mb) CpG islands thought to be involved in changes in DNA methylation (methylation quantitative trait loci [mQTL]). Each functionally connected GWAS-eQTL-mQTL association is referred to as a triplet.
The next arm of the study investigatedthe connections and pathways within each triplet. Three possible scenarios were explored for birthweight GWAS SNPs using a causal interference test (CIT):
- the SNP alters placental DNA methylation, which then influences gene expression
- the SNP first alters placental DNA expression, which then influences methylation
- the SNP influences placental DNA expression and methylation independently, with no notable crossover between their pathways.
Triplets were investigated using the Mendelian randomization (MR) Steiger directionality test to validate the directionality of the pathways found by CIT. Lastly, the possibility of linkage disequilibrium was also studied using the moloc test.
Results
Using CIT, a causal relationship was predicted in 88 of 197 triplets, in which 84 (95.5%) indicated DNA methylation influences gene expression, and 4 (4.5%) indicated gene expression influences DNA methylation. The authors also used the MR Steiger test to investigate triplets to identify possible causal pathways. Using the MR Steiger test, only 3 of 45 (7%) triplets were found to have independent gene expression and methylation pathways. Thirty-eight of 45 (84%) triplets indicated that gene expression influences DNA methylation, and 7 (15%)triplets demonstrated that DNA methylation influences gene expression. Consistent predictions between CIT and the MR Steiger test revealed 3 triplets in which DNA methylation influences gene expression for the following genes: WNT3A, CTDNEP1, and RANBP2. Additionally, a strong colocalization signal was found among birthweight, DNA methylation, and gene expression for the following genes: PLEKHA1, FES, PRMT7, and CTDNEP1. Gene set enrichment analysis was performed as well and found that low birthweight is associated in substantial upregulation of genes associated with oxidative stress, immune response, adipogenesis, myogenesis, and the production of pancreatic ß cells.
Study strengths and limitations
The study is one of the first to identify regulatory targets for placental DNA methylation and gene expression in previously identified GWAS loci associated with low birthweight. For example, DNA methylation was found to influence gene expression of WNT3A, CTDNEP1, and RANBP2, which have previously been shown in animal studies to impact the vascularization and development of the placenta, embryogenesis, and fetal growth. The study also identified 4 genes (PLEKHA1, FES, PRMT7, and CTDNEP1) thought to have direct regulatory influence on placental DNA methylation and gene expression.
A limitation of the study is that it could not distinguish between whether the epigenetic changes we outlined have a maternal or fetal origin. Another limitation is that tissue used by the authors for analysis was a small placental biopsy, which does not accurately reflect the genetic heterogeneity of the placenta. Finally, this study does not establish causality between the identified epigenetic pathways and low birthweight. ●
We know that the placenta is critical to in utero development. This study begins to explore the genetic changes and programming in the placenta that may have profound effects on health and well-being both early and later in life.
- Li LS, Li DZ. A genetic approach to the etiologic investigation of isolated intrauterine growth restriction. Am J Obstet Gynecol. 2021;225:695-696. doi: 10.1016/j.ajog.2021 .07.021.
- Borrell A, Grande M, Pauta M, et al. Chromosomal microarray analysis in fetuses with growth restriction and normal karyotype: a systematic review and meta-analysis. Fetal Diagn Ther. 2018;44:1-9. doi: 10.1159/000479506.
Whole exome sequencing’s role in diagnosing genetic causes of FGR with and without associated anomalies
Mone F, Mellis R, Gabriel H, et al. Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am J Obstet Gynecol. Published online October 7, 2022. doi:10.1016/j.ajog.2022.09.045
Multiple factors can play a role in FGR, including inherent maternal, placental, or fetal factors; the environment; and/or nutrition. However, prenatal diagnosis is an important consideration when exploring the underlying etiology for a growth-restricted fetus, especially in severe or early-onset cases. Many genetic conditions do not result in structural anomalies but can disrupt overall growth. Additionally, phenotyping in the prenatal period is limited and can miss more subtle physical differences that could point to a genetic cause.
When compared with karyotype, chromosomal microarray (CMA) has been shown to increase the diagnostic yield in cases of isolated early FGR by 5%,1,2 and the incidence of chromosomal abnormalities has been reported to be as high as 19% in this population. Let’s explore the data on exome sequencing for prenatal diagnosis in cases of isolated FGR.
Meta-analysis details
In this meta-analysis, the authors reviewed 19 cohort studies or case series that investigated the yield of prenatal sequencing in fetuses with intrauterine growth restriction (IUGR) or short long bones, both in association with and without additional anomalies. All cases had nondiagnostic cytogenetic results. Fetal DNA in most cases was obtained through amniocentesis. Variants classified as likely pathogenic and pathogenic were considered diagnostic. The authors then calculated the incremental yield of prenatal sequencing over cytogenetic studies as a pooled value, comparing the following groups:
- isolated FGR
- growth restriction with associated anomalies
- isolated short long bones
- short long bones with additional skeletal features.
Study outcomes
The total number of cases were as follows: isolated IUGR (n = 71), IUGR associated with additional anomalies (n = 45), isolated short long bones (n = 84), and short long bones associated with additional skeletal findings (n = 252). Causative pathogenic or likely pathogenic variants were identified in 224 (50%) cases. Apparent incremental yields with prenatal sequencing were as follows for the 4 groups (as illustrated in the FIGURE):

- 4% in isolated IUGR (95% confidence interval [CI], -5%–12%)
- 30% in IUGR with additional anomalies (95% CI, 13%–47%)
- 48% in isolated short long bones (95% CI, 26%–70%)
- 68% in short long bones with additional skeletal changes (95% CI, 58%–77%).
Overall, the authors concluded that prenatal sequencing does not improve prenatal diagnosis in cases of isolated IUGR. The majority of these cases were thought to be related to placental insufficiency.
Strengths and limitations
The main limitation of this study with regard to our discussion is the small study populationof isolated growth restriction. The authors indicate that the number of cases of isolated IUGR were too small to draw firm conclusions. Another limitation was the heterogeneity of the isolated FGR population, which was not limited to severe or early-onset cases. However, the authors did demonstrate that growth restriction in association with fetal anomalies has very high genetic yield rates with prenatal sequencing.
Not surprisingly, there is a high yield of diagnosing genetic conditions in pregnancies complicated by isolated or nonisolated short long bones or in cases of growth restriction with multisystem abnormalities. Based on the results of this study, the authors advise against sending for exome sequencing in cases of isolated growth restriction with coexisting evidence of placental insufficiency.
Continue to: Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?...
Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?
Zhou H, Fu F, Wang Y, et al. Genetic causes of isolated and severe fetal growth restriction in normal chromosomal microarray analysis. Int J Gynaecol Obstet. Published online December 10, 2022. doi:10.1002/ijgo.14620
Severe FGR is diagnosed based on an estimated fetal weight (EFW) or abdominal circumference (AC) below the third percentile. As we discussed in the above study by Mone and colleagues, it does not appear that prenatal sequencing significantly improves the diagnostic yield in all isolated FGR cases. However, this has not been previously explored in isolated severe FGR or cases of early-onset FGR (<32 weeks’ gestation). We know that several monogenic conditions are associated with severe and early-onset isolated fetal growth impairment, including but not limited to Cornelia de Lange syndrome, Smith-Lemli-Opitz syndrome, and Meier-Gorlin syndrome. Often, these syndromes can present in the prenatal period without other phenotypic findings. Therefore, this study explored the possibility that prenatal sequencing plays an important role for severe cases of FGR with nondiagnostic CMA and/or karyotype.
Retrospective study details
Zhou and colleagues retrospectively analyzed 51 cases of severe (EFW or AC below the third percentile) isolated FGR with negative CMA who underwent trio whole exome sequencing, which includes submitting fetal cells as well as both parental samples for testing. Patients with abnormal toxoplasmosis, rubella, cytomegalovirus, and herpes simplex virus (TORCH) tests; structural anomalies; and multiple gestation were excluded from the analysis. As in the study by Mone et al, variants classified as likely pathogenic and pathogenic were categorized as diagnostic.
Results
Eight of 51 cases (15.7%) with severe isolated FGR had diagnostic findings on trio whole exome sequencing as shown in the TABLE. Another 8 cases (15.7%) were found to have variants of unknown significance, of which 2 were later determined to be novel pathogenic variants. Genetic conditions uncovered in this cohort include Cornelia de Lange syndrome, pyruvate dehydrogenase deficiency, Dent disease, trichohepaticenteric syndrome, achondroplasia, osteogenesis imperfecta, Pendred syndrome, and both autosomal dominant type 3A and autosomal recessive type 1A deafness. All 10 cases with diagnostic whole exome sequencing or identified novel pathogenic variants were affected by early-onset FGR (<32 weeks’ gestation). Of these 10 cases, 7 patients underwent pregnancy termination.

To summarize, a total of 10 cases (19.6%) of severe isolated early-onset FGR with negative cytogenetic studies were subsequently diagnosed with an underlying genetic condition using prenatal trio whole exome sequencing.
Strengths and limitations
This study is retrospective and has a small sample size (n = 51) that was mostly limited to early-onset isolated severe FGR. However, the diagnostic yield (19.6%) of whole exome sequencing after negative CMA testing was noteworthy and shows that monogenic conditions are an important consideration in the evaluation of severe early-onset FGR, even in the absence of structural abnormalities.
As indications for exome sequencing during pregnancy continue to evolve, severe isolated FGR is emerging as a high-yield condition in which a subset of patients may benefit from the described testing strategy. We learned from our look at the prior study (Mone et al) that unselected isolated growth restriction with evident placental insufficiency may not benefit from exome sequencing, but this study differs in its selection of early-onset, severe cases—defined by diagnosis before 32 weeks’ gestation and an EFW or AC below the third percentile. Almost 20% of cases who met the aforementioned criteria received a genetic diagnosis from exome sequencing. We should remember to offer genetic counseling and diagnostic testing to our patients with severe growth restriction, even in the absence of additional structural anomalies.
Could epigenetic mechanisms of placental dysregulation explain low birthweight and future cardiometabolic disease?
Tekola-Ayele F, Zeng X, Chatterjee S, et al. Placental multi-omics integration identifies candidate functional genes for birthweight. Nat Commun. 2022;13:2384.
FGR has been linked to greater mortality in childhood and increased risk for cardiometabolic disease in adulthood. While genomewide associations studies (GWAS) have defined areas of interest linking genetic variants to low birthweight, their relationship to epigenetic changes in the placenta as well as biologic and functional mechanisms are not yet well understood.
Multiomics used to identify candidate functional genes for birthweight
This study analyzed the methylation and gene expression patterns of 291 placental samples, integrating findings into pathways of previously defined GWAS variants. Patient samples were obtained from participants in the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Fetal Growth Studies–Singleton cohort. The cohort is ethnically diverse, with 97 Hispanic, 74 White, 71 Black, and 49 Asian participants. Of 286 single nucleotide polymorphisms (SNPs) found to be associated with birthweight, 273 were analyzed as part of the authors’ data set. These were found to have 7,901 unique protein-coding mRNAs (expression quantitative trait loci [eQTL]) and more than 100,000 nearby (within 1 Mb) CpG islands thought to be involved in changes in DNA methylation (methylation quantitative trait loci [mQTL]). Each functionally connected GWAS-eQTL-mQTL association is referred to as a triplet.
The next arm of the study investigatedthe connections and pathways within each triplet. Three possible scenarios were explored for birthweight GWAS SNPs using a causal interference test (CIT):
- the SNP alters placental DNA methylation, which then influences gene expression
- the SNP first alters placental DNA expression, which then influences methylation
- the SNP influences placental DNA expression and methylation independently, with no notable crossover between their pathways.
Triplets were investigated using the Mendelian randomization (MR) Steiger directionality test to validate the directionality of the pathways found by CIT. Lastly, the possibility of linkage disequilibrium was also studied using the moloc test.
Results
Using CIT, a causal relationship was predicted in 88 of 197 triplets, in which 84 (95.5%) indicated DNA methylation influences gene expression, and 4 (4.5%) indicated gene expression influences DNA methylation. The authors also used the MR Steiger test to investigate triplets to identify possible causal pathways. Using the MR Steiger test, only 3 of 45 (7%) triplets were found to have independent gene expression and methylation pathways. Thirty-eight of 45 (84%) triplets indicated that gene expression influences DNA methylation, and 7 (15%)triplets demonstrated that DNA methylation influences gene expression. Consistent predictions between CIT and the MR Steiger test revealed 3 triplets in which DNA methylation influences gene expression for the following genes: WNT3A, CTDNEP1, and RANBP2. Additionally, a strong colocalization signal was found among birthweight, DNA methylation, and gene expression for the following genes: PLEKHA1, FES, PRMT7, and CTDNEP1. Gene set enrichment analysis was performed as well and found that low birthweight is associated in substantial upregulation of genes associated with oxidative stress, immune response, adipogenesis, myogenesis, and the production of pancreatic ß cells.
Study strengths and limitations
The study is one of the first to identify regulatory targets for placental DNA methylation and gene expression in previously identified GWAS loci associated with low birthweight. For example, DNA methylation was found to influence gene expression of WNT3A, CTDNEP1, and RANBP2, which have previously been shown in animal studies to impact the vascularization and development of the placenta, embryogenesis, and fetal growth. The study also identified 4 genes (PLEKHA1, FES, PRMT7, and CTDNEP1) thought to have direct regulatory influence on placental DNA methylation and gene expression.
A limitation of the study is that it could not distinguish between whether the epigenetic changes we outlined have a maternal or fetal origin. Another limitation is that tissue used by the authors for analysis was a small placental biopsy, which does not accurately reflect the genetic heterogeneity of the placenta. Finally, this study does not establish causality between the identified epigenetic pathways and low birthweight. ●
We know that the placenta is critical to in utero development. This study begins to explore the genetic changes and programming in the placenta that may have profound effects on health and well-being both early and later in life.
Whole exome sequencing’s role in diagnosing genetic causes of FGR with and without associated anomalies
Mone F, Mellis R, Gabriel H, et al. Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am J Obstet Gynecol. Published online October 7, 2022. doi:10.1016/j.ajog.2022.09.045
Multiple factors can play a role in FGR, including inherent maternal, placental, or fetal factors; the environment; and/or nutrition. However, prenatal diagnosis is an important consideration when exploring the underlying etiology for a growth-restricted fetus, especially in severe or early-onset cases. Many genetic conditions do not result in structural anomalies but can disrupt overall growth. Additionally, phenotyping in the prenatal period is limited and can miss more subtle physical differences that could point to a genetic cause.
When compared with karyotype, chromosomal microarray (CMA) has been shown to increase the diagnostic yield in cases of isolated early FGR by 5%,1,2 and the incidence of chromosomal abnormalities has been reported to be as high as 19% in this population. Let’s explore the data on exome sequencing for prenatal diagnosis in cases of isolated FGR.
Meta-analysis details
In this meta-analysis, the authors reviewed 19 cohort studies or case series that investigated the yield of prenatal sequencing in fetuses with intrauterine growth restriction (IUGR) or short long bones, both in association with and without additional anomalies. All cases had nondiagnostic cytogenetic results. Fetal DNA in most cases was obtained through amniocentesis. Variants classified as likely pathogenic and pathogenic were considered diagnostic. The authors then calculated the incremental yield of prenatal sequencing over cytogenetic studies as a pooled value, comparing the following groups:
- isolated FGR
- growth restriction with associated anomalies
- isolated short long bones
- short long bones with additional skeletal features.
Study outcomes
The total number of cases were as follows: isolated IUGR (n = 71), IUGR associated with additional anomalies (n = 45), isolated short long bones (n = 84), and short long bones associated with additional skeletal findings (n = 252). Causative pathogenic or likely pathogenic variants were identified in 224 (50%) cases. Apparent incremental yields with prenatal sequencing were as follows for the 4 groups (as illustrated in the FIGURE):

- 4% in isolated IUGR (95% confidence interval [CI], -5%–12%)
- 30% in IUGR with additional anomalies (95% CI, 13%–47%)
- 48% in isolated short long bones (95% CI, 26%–70%)
- 68% in short long bones with additional skeletal changes (95% CI, 58%–77%).
Overall, the authors concluded that prenatal sequencing does not improve prenatal diagnosis in cases of isolated IUGR. The majority of these cases were thought to be related to placental insufficiency.
Strengths and limitations
The main limitation of this study with regard to our discussion is the small study populationof isolated growth restriction. The authors indicate that the number of cases of isolated IUGR were too small to draw firm conclusions. Another limitation was the heterogeneity of the isolated FGR population, which was not limited to severe or early-onset cases. However, the authors did demonstrate that growth restriction in association with fetal anomalies has very high genetic yield rates with prenatal sequencing.
Not surprisingly, there is a high yield of diagnosing genetic conditions in pregnancies complicated by isolated or nonisolated short long bones or in cases of growth restriction with multisystem abnormalities. Based on the results of this study, the authors advise against sending for exome sequencing in cases of isolated growth restriction with coexisting evidence of placental insufficiency.
Continue to: Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?...
Can whole exome sequencing diagnose genetic causes in cases of severe isolated FGR?
Zhou H, Fu F, Wang Y, et al. Genetic causes of isolated and severe fetal growth restriction in normal chromosomal microarray analysis. Int J Gynaecol Obstet. Published online December 10, 2022. doi:10.1002/ijgo.14620
Severe FGR is diagnosed based on an estimated fetal weight (EFW) or abdominal circumference (AC) below the third percentile. As we discussed in the above study by Mone and colleagues, it does not appear that prenatal sequencing significantly improves the diagnostic yield in all isolated FGR cases. However, this has not been previously explored in isolated severe FGR or cases of early-onset FGR (<32 weeks’ gestation). We know that several monogenic conditions are associated with severe and early-onset isolated fetal growth impairment, including but not limited to Cornelia de Lange syndrome, Smith-Lemli-Opitz syndrome, and Meier-Gorlin syndrome. Often, these syndromes can present in the prenatal period without other phenotypic findings. Therefore, this study explored the possibility that prenatal sequencing plays an important role for severe cases of FGR with nondiagnostic CMA and/or karyotype.
Retrospective study details
Zhou and colleagues retrospectively analyzed 51 cases of severe (EFW or AC below the third percentile) isolated FGR with negative CMA who underwent trio whole exome sequencing, which includes submitting fetal cells as well as both parental samples for testing. Patients with abnormal toxoplasmosis, rubella, cytomegalovirus, and herpes simplex virus (TORCH) tests; structural anomalies; and multiple gestation were excluded from the analysis. As in the study by Mone et al, variants classified as likely pathogenic and pathogenic were categorized as diagnostic.
Results
Eight of 51 cases (15.7%) with severe isolated FGR had diagnostic findings on trio whole exome sequencing as shown in the TABLE. Another 8 cases (15.7%) were found to have variants of unknown significance, of which 2 were later determined to be novel pathogenic variants. Genetic conditions uncovered in this cohort include Cornelia de Lange syndrome, pyruvate dehydrogenase deficiency, Dent disease, trichohepaticenteric syndrome, achondroplasia, osteogenesis imperfecta, Pendred syndrome, and both autosomal dominant type 3A and autosomal recessive type 1A deafness. All 10 cases with diagnostic whole exome sequencing or identified novel pathogenic variants were affected by early-onset FGR (<32 weeks’ gestation). Of these 10 cases, 7 patients underwent pregnancy termination.

To summarize, a total of 10 cases (19.6%) of severe isolated early-onset FGR with negative cytogenetic studies were subsequently diagnosed with an underlying genetic condition using prenatal trio whole exome sequencing.
Strengths and limitations
This study is retrospective and has a small sample size (n = 51) that was mostly limited to early-onset isolated severe FGR. However, the diagnostic yield (19.6%) of whole exome sequencing after negative CMA testing was noteworthy and shows that monogenic conditions are an important consideration in the evaluation of severe early-onset FGR, even in the absence of structural abnormalities.
As indications for exome sequencing during pregnancy continue to evolve, severe isolated FGR is emerging as a high-yield condition in which a subset of patients may benefit from the described testing strategy. We learned from our look at the prior study (Mone et al) that unselected isolated growth restriction with evident placental insufficiency may not benefit from exome sequencing, but this study differs in its selection of early-onset, severe cases—defined by diagnosis before 32 weeks’ gestation and an EFW or AC below the third percentile. Almost 20% of cases who met the aforementioned criteria received a genetic diagnosis from exome sequencing. We should remember to offer genetic counseling and diagnostic testing to our patients with severe growth restriction, even in the absence of additional structural anomalies.
Could epigenetic mechanisms of placental dysregulation explain low birthweight and future cardiometabolic disease?
Tekola-Ayele F, Zeng X, Chatterjee S, et al. Placental multi-omics integration identifies candidate functional genes for birthweight. Nat Commun. 2022;13:2384.
FGR has been linked to greater mortality in childhood and increased risk for cardiometabolic disease in adulthood. While genomewide associations studies (GWAS) have defined areas of interest linking genetic variants to low birthweight, their relationship to epigenetic changes in the placenta as well as biologic and functional mechanisms are not yet well understood.
Multiomics used to identify candidate functional genes for birthweight
This study analyzed the methylation and gene expression patterns of 291 placental samples, integrating findings into pathways of previously defined GWAS variants. Patient samples were obtained from participants in the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Fetal Growth Studies–Singleton cohort. The cohort is ethnically diverse, with 97 Hispanic, 74 White, 71 Black, and 49 Asian participants. Of 286 single nucleotide polymorphisms (SNPs) found to be associated with birthweight, 273 were analyzed as part of the authors’ data set. These were found to have 7,901 unique protein-coding mRNAs (expression quantitative trait loci [eQTL]) and more than 100,000 nearby (within 1 Mb) CpG islands thought to be involved in changes in DNA methylation (methylation quantitative trait loci [mQTL]). Each functionally connected GWAS-eQTL-mQTL association is referred to as a triplet.
The next arm of the study investigatedthe connections and pathways within each triplet. Three possible scenarios were explored for birthweight GWAS SNPs using a causal interference test (CIT):
- the SNP alters placental DNA methylation, which then influences gene expression
- the SNP first alters placental DNA expression, which then influences methylation
- the SNP influences placental DNA expression and methylation independently, with no notable crossover between their pathways.
Triplets were investigated using the Mendelian randomization (MR) Steiger directionality test to validate the directionality of the pathways found by CIT. Lastly, the possibility of linkage disequilibrium was also studied using the moloc test.
Results
Using CIT, a causal relationship was predicted in 88 of 197 triplets, in which 84 (95.5%) indicated DNA methylation influences gene expression, and 4 (4.5%) indicated gene expression influences DNA methylation. The authors also used the MR Steiger test to investigate triplets to identify possible causal pathways. Using the MR Steiger test, only 3 of 45 (7%) triplets were found to have independent gene expression and methylation pathways. Thirty-eight of 45 (84%) triplets indicated that gene expression influences DNA methylation, and 7 (15%)triplets demonstrated that DNA methylation influences gene expression. Consistent predictions between CIT and the MR Steiger test revealed 3 triplets in which DNA methylation influences gene expression for the following genes: WNT3A, CTDNEP1, and RANBP2. Additionally, a strong colocalization signal was found among birthweight, DNA methylation, and gene expression for the following genes: PLEKHA1, FES, PRMT7, and CTDNEP1. Gene set enrichment analysis was performed as well and found that low birthweight is associated in substantial upregulation of genes associated with oxidative stress, immune response, adipogenesis, myogenesis, and the production of pancreatic ß cells.
Study strengths and limitations
The study is one of the first to identify regulatory targets for placental DNA methylation and gene expression in previously identified GWAS loci associated with low birthweight. For example, DNA methylation was found to influence gene expression of WNT3A, CTDNEP1, and RANBP2, which have previously been shown in animal studies to impact the vascularization and development of the placenta, embryogenesis, and fetal growth. The study also identified 4 genes (PLEKHA1, FES, PRMT7, and CTDNEP1) thought to have direct regulatory influence on placental DNA methylation and gene expression.
A limitation of the study is that it could not distinguish between whether the epigenetic changes we outlined have a maternal or fetal origin. Another limitation is that tissue used by the authors for analysis was a small placental biopsy, which does not accurately reflect the genetic heterogeneity of the placenta. Finally, this study does not establish causality between the identified epigenetic pathways and low birthweight. ●
We know that the placenta is critical to in utero development. This study begins to explore the genetic changes and programming in the placenta that may have profound effects on health and well-being both early and later in life.
- Li LS, Li DZ. A genetic approach to the etiologic investigation of isolated intrauterine growth restriction. Am J Obstet Gynecol. 2021;225:695-696. doi: 10.1016/j.ajog.2021 .07.021.
- Borrell A, Grande M, Pauta M, et al. Chromosomal microarray analysis in fetuses with growth restriction and normal karyotype: a systematic review and meta-analysis. Fetal Diagn Ther. 2018;44:1-9. doi: 10.1159/000479506.
- Li LS, Li DZ. A genetic approach to the etiologic investigation of isolated intrauterine growth restriction. Am J Obstet Gynecol. 2021;225:695-696. doi: 10.1016/j.ajog.2021 .07.021.
- Borrell A, Grande M, Pauta M, et al. Chromosomal microarray analysis in fetuses with growth restriction and normal karyotype: a systematic review and meta-analysis. Fetal Diagn Ther. 2018;44:1-9. doi: 10.1159/000479506.
