User login
SAN FRANCISCO – A new analysis of cholera strains suggests that bacteriophages – viruses that prey on bacteria – are engaged in an evolutionary arms race with the Vibrio cholerae bacteria, and the dynamic between the two organisms may be an important factor in determining which strain of cholera goes on to cause a pandemic.
The work, presented by Kim Seed, PhD, at IDWeek, an annual scientific meeting on infectious diseases, examined a defense mechanism in V. cholerae, called phage inducible chromosomal island like element (PLE), as well as a unique mechanism in the bacteriophage to counter it. The work adds insight into the cholera strains that could emerge to produce future epidemics, and could even inform the use of bacteriophages as prophylactic agents to counter V. cholerae infection
In her talk, Dr. Seed described the dynamics of the current cholera pandemic, which is the seventh in recorded history and began in the 1960s. Over the past 100 years, six previous strains arose and then vanished, yielding each time to a new strain that became the predominant cholera-causing agent.
“This pattern of evolution, this so-called disappearing act, drives my research – I’m trying to understand what factors promote the evolution of novel genetic variants, and what factors contribute to why those variants disappear,” said Dr. Seed.
That quest brought her to the Bay of Bengal and Bangladesh. Genetic studies have shown this region to be the epicenter of cholera strains. It appears that cholera strains evolve there and then invade other regions of the world as a result of human travel and activity. Go to places in Africa or Asia where there is a cholera outbreak, and you can find cholera bacteria in the water that has the potential to cause human disease – but it won’t be the strain that is causing disease nearby. “(The culprit) is these introduced strains that come from Southeast Asia,” said Dr. Seed.
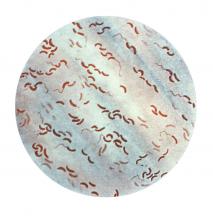
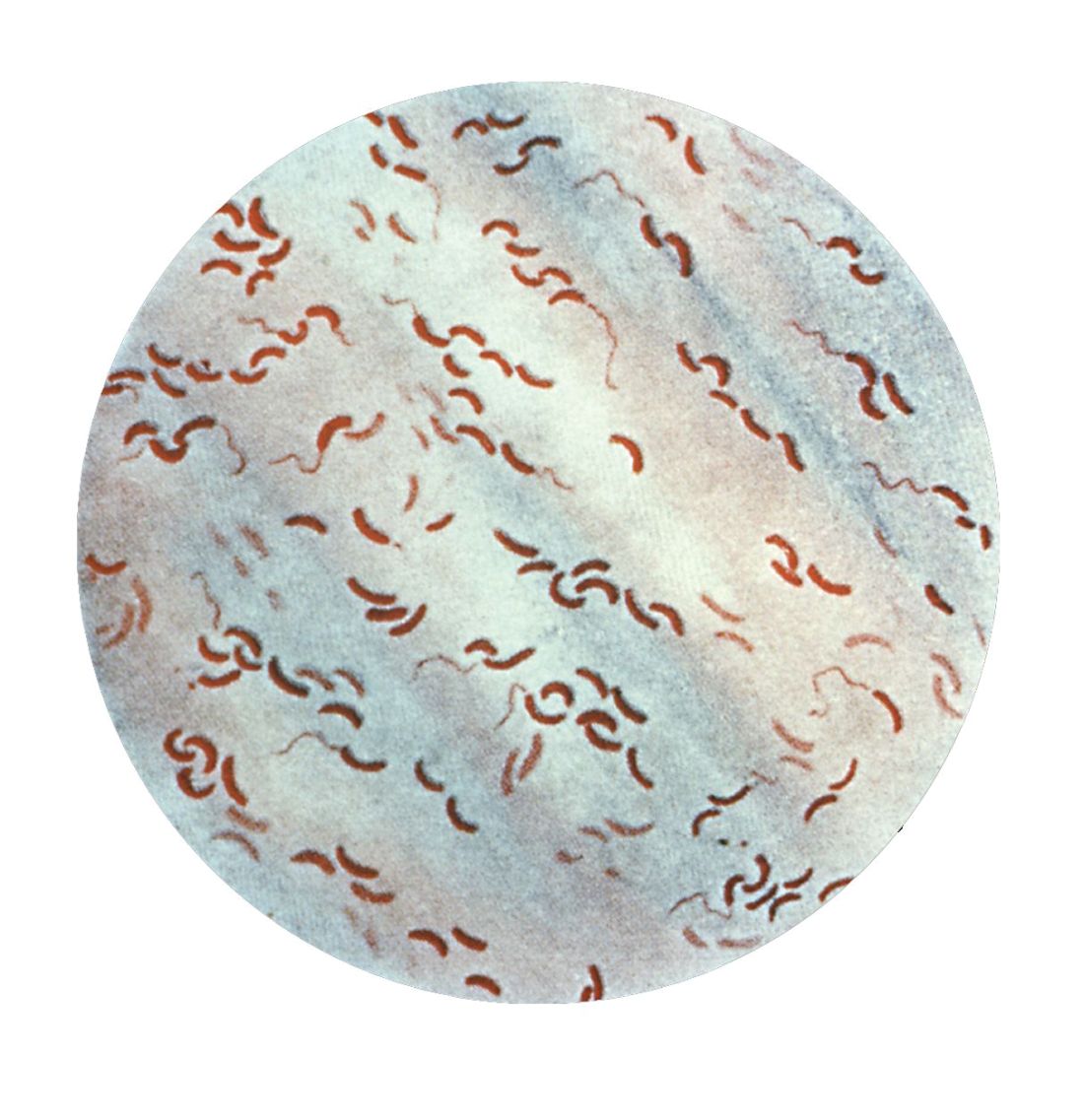
So her team went to Bangladesh, and studied cholera bacteria isolated from patients at the International Centre for Diarrhoeal Disease Research. The current strain is antibiotic resistant, as has been well documented. But Dr. Seed was interested in bacteriophages – viruses that prey on bacteria – because they live in the water supply and can also be isolated from the stool of cholera-infected patients, and it seemed likely that they could be an important selective force.
Indeed, her team found only a few bacteriophages that prey on V. cholerae in the samples from this hospital, and one type predominated in samples collected between 2001 and 2017; a bacteriophage known as ICP1. “This set up a very nice dynamic to be able to study the molecular mechanisms by which co-evolution was occurring in this one specific phage and Vibrio cholerae,” said Dr. Seed.
Genetic analysis revealed a mobile genetic element in V. cholerae – PLE –that conferred specific resistance against ICP1. After an infection, one of the bacteriophage’s proteins leads to excision and transcription of PLE. That produces a predicted 25 proteins, which in turn interfere with ICP1 through an as yet undetermined mechanism. But it’s effective, completely shutting down bacteriophage replication.
That couldn’t be the end of the story, Dr. Seed reasoned. Otherwise the bacteriophage would die out entirely for lack of a vulnerable host. More searching revealed the biggest surprise of all – ICP1, even though it is a virus, contains a complete suite of CRISPR (clustered regularly interspaced short palindromic repeats) apparatus that directly targets the PLE sequence. CRISPR is currently all the rage as a potential tool for genetic modification. It was discovered in bacteria, as a sort of immune response against bacteriophages. The CRISPR DNA contains a guide sequence that is complementary to and binds viral DNA, and then recruits other proteins to destroy the viral blueprint.
But here, for the first and only time, Dr. Seed’s team found that a bacteriophage had turned the tables, somehow capturing a CRISPR system of its own and turning it against its host’s defense system. Soon after infection, PLE switches on in response to its bacteriophage trigger, but the ICP1 counters by activating its CRISPR system, which is effective enough to allow the bacteriophage to reproduce.
The researchers then examined historical samples, and found another surprise: The appearance of CRISPR in ICP1 predated the appearance of the PLE variant that it targeted in V. cholerae. A little more digging revealed older variants of PLE, now gone from the V. cholerae population. “This explains why ICP1 had to have CRISPR, so it could overcome these previously prevalent genetic variants,” said Dr. Seed.
All told, the researchers found five unique PLE variants dating back to 1931, and the co-evolution of V. cholerae and ICP1 no doubt stretch much farther into the dim past. More recently, they found that previous strains of V. cholerae that went extinct also had different variations of PLE, suggesting that it may have been a temporary evolutionary victory by ICP1 over a PLE variant that caused the demise of an existing V. cholerae strain. But each time, it seems the bacteria responded with a new PLE variant, prolonging the arms race.
The work has the potential to affect other bacterial diseases, since most bacteria have phages that prey on them. “I have no doubt that they are a strong presence and selective force on all pathogens. People haven’t done so much work on that yet, but I think it’s coming,” said Dr. Seed.
SOURCE: Seed K. et al. ID Week 2018. Abstract 954.
SAN FRANCISCO – A new analysis of cholera strains suggests that bacteriophages – viruses that prey on bacteria – are engaged in an evolutionary arms race with the Vibrio cholerae bacteria, and the dynamic between the two organisms may be an important factor in determining which strain of cholera goes on to cause a pandemic.
The work, presented by Kim Seed, PhD, at IDWeek, an annual scientific meeting on infectious diseases, examined a defense mechanism in V. cholerae, called phage inducible chromosomal island like element (PLE), as well as a unique mechanism in the bacteriophage to counter it. The work adds insight into the cholera strains that could emerge to produce future epidemics, and could even inform the use of bacteriophages as prophylactic agents to counter V. cholerae infection
In her talk, Dr. Seed described the dynamics of the current cholera pandemic, which is the seventh in recorded history and began in the 1960s. Over the past 100 years, six previous strains arose and then vanished, yielding each time to a new strain that became the predominant cholera-causing agent.
“This pattern of evolution, this so-called disappearing act, drives my research – I’m trying to understand what factors promote the evolution of novel genetic variants, and what factors contribute to why those variants disappear,” said Dr. Seed.
That quest brought her to the Bay of Bengal and Bangladesh. Genetic studies have shown this region to be the epicenter of cholera strains. It appears that cholera strains evolve there and then invade other regions of the world as a result of human travel and activity. Go to places in Africa or Asia where there is a cholera outbreak, and you can find cholera bacteria in the water that has the potential to cause human disease – but it won’t be the strain that is causing disease nearby. “(The culprit) is these introduced strains that come from Southeast Asia,” said Dr. Seed.
So her team went to Bangladesh, and studied cholera bacteria isolated from patients at the International Centre for Diarrhoeal Disease Research. The current strain is antibiotic resistant, as has been well documented. But Dr. Seed was interested in bacteriophages – viruses that prey on bacteria – because they live in the water supply and can also be isolated from the stool of cholera-infected patients, and it seemed likely that they could be an important selective force.
Indeed, her team found only a few bacteriophages that prey on V. cholerae in the samples from this hospital, and one type predominated in samples collected between 2001 and 2017; a bacteriophage known as ICP1. “This set up a very nice dynamic to be able to study the molecular mechanisms by which co-evolution was occurring in this one specific phage and Vibrio cholerae,” said Dr. Seed.
Genetic analysis revealed a mobile genetic element in V. cholerae – PLE –that conferred specific resistance against ICP1. After an infection, one of the bacteriophage’s proteins leads to excision and transcription of PLE. That produces a predicted 25 proteins, which in turn interfere with ICP1 through an as yet undetermined mechanism. But it’s effective, completely shutting down bacteriophage replication.
That couldn’t be the end of the story, Dr. Seed reasoned. Otherwise the bacteriophage would die out entirely for lack of a vulnerable host. More searching revealed the biggest surprise of all – ICP1, even though it is a virus, contains a complete suite of CRISPR (clustered regularly interspaced short palindromic repeats) apparatus that directly targets the PLE sequence. CRISPR is currently all the rage as a potential tool for genetic modification. It was discovered in bacteria, as a sort of immune response against bacteriophages. The CRISPR DNA contains a guide sequence that is complementary to and binds viral DNA, and then recruits other proteins to destroy the viral blueprint.
But here, for the first and only time, Dr. Seed’s team found that a bacteriophage had turned the tables, somehow capturing a CRISPR system of its own and turning it against its host’s defense system. Soon after infection, PLE switches on in response to its bacteriophage trigger, but the ICP1 counters by activating its CRISPR system, which is effective enough to allow the bacteriophage to reproduce.
The researchers then examined historical samples, and found another surprise: The appearance of CRISPR in ICP1 predated the appearance of the PLE variant that it targeted in V. cholerae. A little more digging revealed older variants of PLE, now gone from the V. cholerae population. “This explains why ICP1 had to have CRISPR, so it could overcome these previously prevalent genetic variants,” said Dr. Seed.
All told, the researchers found five unique PLE variants dating back to 1931, and the co-evolution of V. cholerae and ICP1 no doubt stretch much farther into the dim past. More recently, they found that previous strains of V. cholerae that went extinct also had different variations of PLE, suggesting that it may have been a temporary evolutionary victory by ICP1 over a PLE variant that caused the demise of an existing V. cholerae strain. But each time, it seems the bacteria responded with a new PLE variant, prolonging the arms race.
The work has the potential to affect other bacterial diseases, since most bacteria have phages that prey on them. “I have no doubt that they are a strong presence and selective force on all pathogens. People haven’t done so much work on that yet, but I think it’s coming,” said Dr. Seed.
SOURCE: Seed K. et al. ID Week 2018. Abstract 954.
SAN FRANCISCO – A new analysis of cholera strains suggests that bacteriophages – viruses that prey on bacteria – are engaged in an evolutionary arms race with the Vibrio cholerae bacteria, and the dynamic between the two organisms may be an important factor in determining which strain of cholera goes on to cause a pandemic.
The work, presented by Kim Seed, PhD, at IDWeek, an annual scientific meeting on infectious diseases, examined a defense mechanism in V. cholerae, called phage inducible chromosomal island like element (PLE), as well as a unique mechanism in the bacteriophage to counter it. The work adds insight into the cholera strains that could emerge to produce future epidemics, and could even inform the use of bacteriophages as prophylactic agents to counter V. cholerae infection
In her talk, Dr. Seed described the dynamics of the current cholera pandemic, which is the seventh in recorded history and began in the 1960s. Over the past 100 years, six previous strains arose and then vanished, yielding each time to a new strain that became the predominant cholera-causing agent.
“This pattern of evolution, this so-called disappearing act, drives my research – I’m trying to understand what factors promote the evolution of novel genetic variants, and what factors contribute to why those variants disappear,” said Dr. Seed.
That quest brought her to the Bay of Bengal and Bangladesh. Genetic studies have shown this region to be the epicenter of cholera strains. It appears that cholera strains evolve there and then invade other regions of the world as a result of human travel and activity. Go to places in Africa or Asia where there is a cholera outbreak, and you can find cholera bacteria in the water that has the potential to cause human disease – but it won’t be the strain that is causing disease nearby. “(The culprit) is these introduced strains that come from Southeast Asia,” said Dr. Seed.
So her team went to Bangladesh, and studied cholera bacteria isolated from patients at the International Centre for Diarrhoeal Disease Research. The current strain is antibiotic resistant, as has been well documented. But Dr. Seed was interested in bacteriophages – viruses that prey on bacteria – because they live in the water supply and can also be isolated from the stool of cholera-infected patients, and it seemed likely that they could be an important selective force.
Indeed, her team found only a few bacteriophages that prey on V. cholerae in the samples from this hospital, and one type predominated in samples collected between 2001 and 2017; a bacteriophage known as ICP1. “This set up a very nice dynamic to be able to study the molecular mechanisms by which co-evolution was occurring in this one specific phage and Vibrio cholerae,” said Dr. Seed.
Genetic analysis revealed a mobile genetic element in V. cholerae – PLE –that conferred specific resistance against ICP1. After an infection, one of the bacteriophage’s proteins leads to excision and transcription of PLE. That produces a predicted 25 proteins, which in turn interfere with ICP1 through an as yet undetermined mechanism. But it’s effective, completely shutting down bacteriophage replication.
That couldn’t be the end of the story, Dr. Seed reasoned. Otherwise the bacteriophage would die out entirely for lack of a vulnerable host. More searching revealed the biggest surprise of all – ICP1, even though it is a virus, contains a complete suite of CRISPR (clustered regularly interspaced short palindromic repeats) apparatus that directly targets the PLE sequence. CRISPR is currently all the rage as a potential tool for genetic modification. It was discovered in bacteria, as a sort of immune response against bacteriophages. The CRISPR DNA contains a guide sequence that is complementary to and binds viral DNA, and then recruits other proteins to destroy the viral blueprint.
But here, for the first and only time, Dr. Seed’s team found that a bacteriophage had turned the tables, somehow capturing a CRISPR system of its own and turning it against its host’s defense system. Soon after infection, PLE switches on in response to its bacteriophage trigger, but the ICP1 counters by activating its CRISPR system, which is effective enough to allow the bacteriophage to reproduce.
The researchers then examined historical samples, and found another surprise: The appearance of CRISPR in ICP1 predated the appearance of the PLE variant that it targeted in V. cholerae. A little more digging revealed older variants of PLE, now gone from the V. cholerae population. “This explains why ICP1 had to have CRISPR, so it could overcome these previously prevalent genetic variants,” said Dr. Seed.
All told, the researchers found five unique PLE variants dating back to 1931, and the co-evolution of V. cholerae and ICP1 no doubt stretch much farther into the dim past. More recently, they found that previous strains of V. cholerae that went extinct also had different variations of PLE, suggesting that it may have been a temporary evolutionary victory by ICP1 over a PLE variant that caused the demise of an existing V. cholerae strain. But each time, it seems the bacteria responded with a new PLE variant, prolonging the arms race.
The work has the potential to affect other bacterial diseases, since most bacteria have phages that prey on them. “I have no doubt that they are a strong presence and selective force on all pathogens. People haven’t done so much work on that yet, but I think it’s coming,” said Dr. Seed.
SOURCE: Seed K. et al. ID Week 2018. Abstract 954.
REPORTING FROM IDWEEK 2018
Key clinical point: Bacteriophages place selective pressure on bacteria that may influence the emergence of new pathogenic strains.
Major finding: Bacteriophages turn on their own CRISPR system to target cholera defensive genes.
Study details: Genetic analysis of an antibiotic resistant strain and attacking bacteriophage strains.
Disclosures: The National Institutes of Health and the Chan Zuckerberg Biohub provided research funding.
Source: Seed K et al. ID Week 2018. Abstract 954.