User login
Traditionally, a hereditary colorectal cancer syndrome (HCCS) was suspected in individuals with an obvious personal and/or family cancer phenotype informed by a three-generation family cancer history. Family history is still required to inform cancer risk. Documentation of age at cancer diagnosis, age of relatives’ deaths, and key intestinal and extraintestinal features of a HCCS (for example, macrocephaly, café au lait spots, polyp number, size, and histology) are requisite. Historically, Sanger sequencing was used to determine the presence of a suspected single pathogenic germline variant (PGV). If no PGV was detected, another PGV would be sought. This old “single gene/single syndrome” testing was expensive, time consuming, and inefficient, and has been supplanted by multigene cancer panel testing (MGPT). MGPT-driven low-cost, high-throughput testing has widespread insurance coverage in eligible patients. Since considerable clinical phenotypic overlap exists between HCCSs, casting a broader net for determining PGV, compared with a more limited approach, allows for greater identification of carriers of PGV as well as variants of uncertain significance.
The frequency of PGV detection by MGPT in individuals with CRC is dependent on age at diagnosis and presence of DNA mismatch repair (MMR) deficiency in the tumor. According to one review, PGVs on MGPT are detected in approximately 10% and 34% of individuals aged more than 50 and more than 35 years, respectively.1 Pearlman and colleagues performed MGPT in 450 patients with CRC less than 50 years.2 PGV were found in 8% and 83.3% of cases with MMR-proficient and -deficient tumors, respectively. Overall, 33.3% of patients did not meet genetic testing criteria for the gene in which a PGV was detected, raising the impetus to consider MGPT in all patients with CRC. The Collaborative Group of the Americas on Inherited Gastrointestinal Cancer and National Comprehensive Cancer Network provide guidance on who warrants PGV testing.3,4
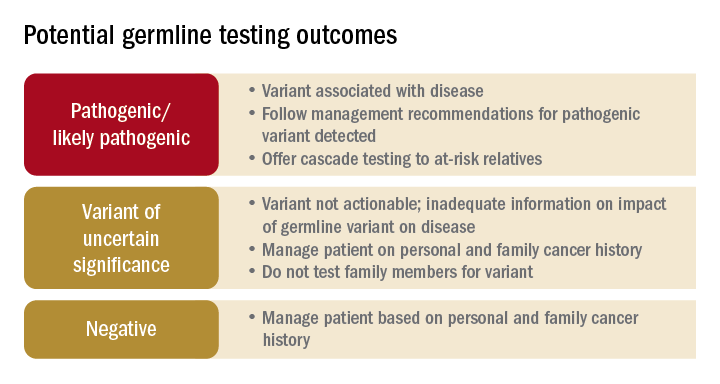
Germline testing outcomes and general approaches to patient management are provided in the graphic. HCCS are common and MGPT has broadened the identification of carriers of PGVs. In spite of advances in genetic testing technology, family history remains crucial to deploying risk-mitigation measures, regardless of the results of genetic testing.
Dr. Burke is in the department of gastroenterology, hepatology, and nutrition at the Cleveland Clinic. She disclosed ties to Janssen Pharma, Emtora Biosciences, Freenome, SLA Pharma, and Ambry Genetics. Dr. Burke is a member of the U.S. Multi-Society Task Force on Colorectal Cancer, National Comprehensive Cancer Network Guideline on Genetic/Familial High-Risk Assessment: Colorectal. These remarks were made during one of the AGA Postgraduate Course sessions held at DDW 2022.
References
1. Stoffel E and Murphy CC. Gastroenterology. 2020 Jan;158(2):341-353.
2. Pearlman R et al. JAMA Oncol. 2017 Apr 1;3(4):464-471.
3. Heald B et al. Fam Cancer. 2020 Jul;19(3):223-239.
4. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal Version 1.2022. 2022 Jun 8.
Traditionally, a hereditary colorectal cancer syndrome (HCCS) was suspected in individuals with an obvious personal and/or family cancer phenotype informed by a three-generation family cancer history. Family history is still required to inform cancer risk. Documentation of age at cancer diagnosis, age of relatives’ deaths, and key intestinal and extraintestinal features of a HCCS (for example, macrocephaly, café au lait spots, polyp number, size, and histology) are requisite. Historically, Sanger sequencing was used to determine the presence of a suspected single pathogenic germline variant (PGV). If no PGV was detected, another PGV would be sought. This old “single gene/single syndrome” testing was expensive, time consuming, and inefficient, and has been supplanted by multigene cancer panel testing (MGPT). MGPT-driven low-cost, high-throughput testing has widespread insurance coverage in eligible patients. Since considerable clinical phenotypic overlap exists between HCCSs, casting a broader net for determining PGV, compared with a more limited approach, allows for greater identification of carriers of PGV as well as variants of uncertain significance.

The frequency of PGV detection by MGPT in individuals with CRC is dependent on age at diagnosis and presence of DNA mismatch repair (MMR) deficiency in the tumor. According to one review, PGVs on MGPT are detected in approximately 10% and 34% of individuals aged more than 50 and more than 35 years, respectively.1 Pearlman and colleagues performed MGPT in 450 patients with CRC less than 50 years.2 PGV were found in 8% and 83.3% of cases with MMR-proficient and -deficient tumors, respectively. Overall, 33.3% of patients did not meet genetic testing criteria for the gene in which a PGV was detected, raising the impetus to consider MGPT in all patients with CRC. The Collaborative Group of the Americas on Inherited Gastrointestinal Cancer and National Comprehensive Cancer Network provide guidance on who warrants PGV testing.3,4
Germline testing outcomes and general approaches to patient management are provided in the graphic. HCCS are common and MGPT has broadened the identification of carriers of PGVs. In spite of advances in genetic testing technology, family history remains crucial to deploying risk-mitigation measures, regardless of the results of genetic testing.
Dr. Burke is in the department of gastroenterology, hepatology, and nutrition at the Cleveland Clinic. She disclosed ties to Janssen Pharma, Emtora Biosciences, Freenome, SLA Pharma, and Ambry Genetics. Dr. Burke is a member of the U.S. Multi-Society Task Force on Colorectal Cancer, National Comprehensive Cancer Network Guideline on Genetic/Familial High-Risk Assessment: Colorectal. These remarks were made during one of the AGA Postgraduate Course sessions held at DDW 2022.
References
1. Stoffel E and Murphy CC. Gastroenterology. 2020 Jan;158(2):341-353.
2. Pearlman R et al. JAMA Oncol. 2017 Apr 1;3(4):464-471.
3. Heald B et al. Fam Cancer. 2020 Jul;19(3):223-239.
4. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal Version 1.2022. 2022 Jun 8.
Traditionally, a hereditary colorectal cancer syndrome (HCCS) was suspected in individuals with an obvious personal and/or family cancer phenotype informed by a three-generation family cancer history. Family history is still required to inform cancer risk. Documentation of age at cancer diagnosis, age of relatives’ deaths, and key intestinal and extraintestinal features of a HCCS (for example, macrocephaly, café au lait spots, polyp number, size, and histology) are requisite. Historically, Sanger sequencing was used to determine the presence of a suspected single pathogenic germline variant (PGV). If no PGV was detected, another PGV would be sought. This old “single gene/single syndrome” testing was expensive, time consuming, and inefficient, and has been supplanted by multigene cancer panel testing (MGPT). MGPT-driven low-cost, high-throughput testing has widespread insurance coverage in eligible patients. Since considerable clinical phenotypic overlap exists between HCCSs, casting a broader net for determining PGV, compared with a more limited approach, allows for greater identification of carriers of PGV as well as variants of uncertain significance.

The frequency of PGV detection by MGPT in individuals with CRC is dependent on age at diagnosis and presence of DNA mismatch repair (MMR) deficiency in the tumor. According to one review, PGVs on MGPT are detected in approximately 10% and 34% of individuals aged more than 50 and more than 35 years, respectively.1 Pearlman and colleagues performed MGPT in 450 patients with CRC less than 50 years.2 PGV were found in 8% and 83.3% of cases with MMR-proficient and -deficient tumors, respectively. Overall, 33.3% of patients did not meet genetic testing criteria for the gene in which a PGV was detected, raising the impetus to consider MGPT in all patients with CRC. The Collaborative Group of the Americas on Inherited Gastrointestinal Cancer and National Comprehensive Cancer Network provide guidance on who warrants PGV testing.3,4
Germline testing outcomes and general approaches to patient management are provided in the graphic. HCCS are common and MGPT has broadened the identification of carriers of PGVs. In spite of advances in genetic testing technology, family history remains crucial to deploying risk-mitigation measures, regardless of the results of genetic testing.
Dr. Burke is in the department of gastroenterology, hepatology, and nutrition at the Cleveland Clinic. She disclosed ties to Janssen Pharma, Emtora Biosciences, Freenome, SLA Pharma, and Ambry Genetics. Dr. Burke is a member of the U.S. Multi-Society Task Force on Colorectal Cancer, National Comprehensive Cancer Network Guideline on Genetic/Familial High-Risk Assessment: Colorectal. These remarks were made during one of the AGA Postgraduate Course sessions held at DDW 2022.
References
1. Stoffel E and Murphy CC. Gastroenterology. 2020 Jan;158(2):341-353.
2. Pearlman R et al. JAMA Oncol. 2017 Apr 1;3(4):464-471.
3. Heald B et al. Fam Cancer. 2020 Jul;19(3):223-239.
4. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal Version 1.2022. 2022 Jun 8.
AT DDW 2022
